
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,590,844 – 20,590,965 |
| Length | 121 |
| Max. P | 0.853138 |

| Location | 20,590,844 – 20,590,946 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -22.96 |
| Energy contribution | -22.68 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843083 |
| Prediction | RNA |
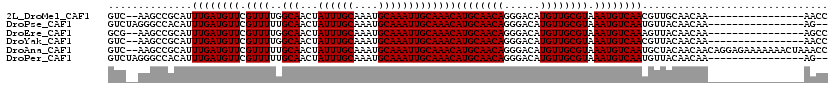
Download alignment: ClustalW | MAF
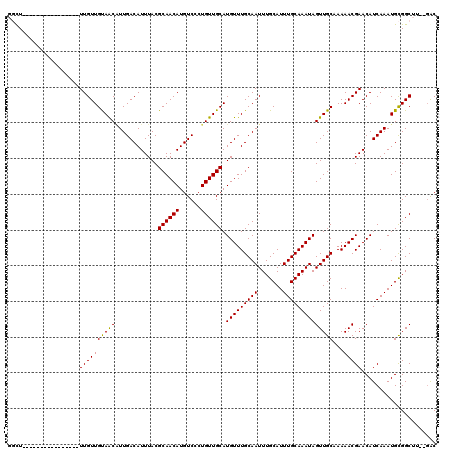
>2L_DroMel_CAF1 20590844 102 + 22407834 GGUU----------------UUGUUGCAACGUUGACAUUUACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCCAAAACGAACAUCAAAUGCGGCUU--GAC .(((----------------(((..(((((............((((((......)))))).(((((((((.......)))))))))))))))))))).....((((.......))--)). ( -28.20) >DroPse_CAF1 154578 102 + 1 --CU----------------UUGUUGUAACAUUGACAUUUACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCAAAAACGAACAUCAAAUGUGGCCCUAGAC --.(----------------((((((((((............((((((......)))))).(((((((((.......)))))))))))))))...))))).................... ( -23.80) >DroEre_CAF1 95927 102 + 1 GGCU----------------UUGUUGUAACUUUGACAUUUACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCCAAAACGAACAUCAAAUGCGGCUU--CGC .((.----------------..((((((..(((((..(((..((((((......)))))).(((((((((.......)))))))))(((.....))))))..)))))))))))..--.)) ( -24.80) >DroYak_CAF1 99639 102 + 1 GGUU----------------UUGUUGUAACGUUGACAUUUACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCCAAAACGAACAUCAAAUGCGGCUU--GAC .(((----------------(((..(((((............((((((......)))))).(((((((((.......)))))))))))))))))))).....((((.......))--)). ( -26.20) >DroAna_CAF1 99022 118 + 1 GGUUUAGUUUUUUUCUCCUGUUGUUGUAGCAUUGACAUUUACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCAAAAACGAACAUCAAAUGCGGCUU--GAC .(((.((((..........(((.(((((((............((((((......)))))).(((((((((.......)))))))))))))))).)))(.....)......)))).--))) ( -26.80) >DroPer_CAF1 160703 102 + 1 --CU----------------UUGUUGUAACAUUGACAUUUACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCAAAAACGAACAUCAAAUGUGGCCCUAGAC --.(----------------((((((((((............((((((......)))))).(((((((((.......)))))))))))))))...))))).................... ( -23.80) >consensus GGCU________________UUGUUGUAACAUUGACAUUUACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCAAAAACGAACAUCAAAUGCGGCUU__GAC ....................((((((((((............((((((......)))))).(((((((((.......))))))))))))))...)))))..................... (-22.96 = -22.68 + -0.28)


| Location | 20,590,844 – 20,590,946 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
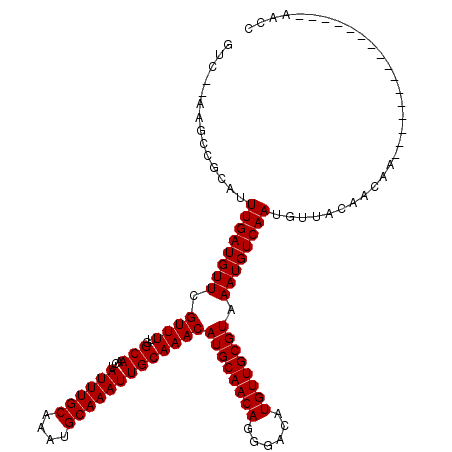
>2L_DroMel_CAF1 20590844 102 - 22407834 GUC--AAGCCGCAUUUGAUGUUCGUUUUGGCAACUAUUUGCAAAUGCAAAUUGCAAACAUGCAACAGGGACAUGUUGCGUAAAUGUCAACGUUGCAACAA----------------AACC (((--(((.....))))))....(((((((((...((((((....))))))))).....((((((...(((((.........)))))...)))))).)))----------------))). ( -28.20) >DroPse_CAF1 154578 102 - 1 GUCUAGGGCCACAUUUGAUGUUCGUUUUUGCAACUAUUUGCAAAUGCAAAUUGCAAACAUGCAACAGGGACAUGUUGCGUAAAUGUCAAUGUUACAACAA----------------AG-- ..........(((((.((((((....((((((...((((((....)))))))))))).((((((((......)))))))).)))))))))))........----------------..-- ( -25.30) >DroEre_CAF1 95927 102 - 1 GCG--AAGCCGCAUUUGAUGUUCGUUUUGGCAACUAUUUGCAAAUGCAAAUUGCAAACAUGCAACAGGGACAUGUUGCGUAAAUGUCAAAGUUACAACAA----------------AGCC (((--....))).((((.(((...((((((((....(((((((.......))))))).((((((((......))))))))...))))))))..))).)))----------------)... ( -29.40) >DroYak_CAF1 99639 102 - 1 GUC--AAGCCGCAUUUGAUGUUCGUUUUGGCAACUAUUUGCAAAUGCAAAUUGCAAACAUGCAACAGGGACAUGUUGCGUAAAUGUCAACGUUACAACAA----------------AACC ...--........((((.(((.(((..(((((....(((((((.......))))))).((((((((......))))))))...))))))))..))).)))----------------)... ( -25.40) >DroAna_CAF1 99022 118 - 1 GUC--AAGCCGCAUUUGAUGUUCGUUUUUGCAACUAUUUGCAAAUGCAAAUUGCAAACAUGCAACAGGGACAUGUUGCGUAAAUGUCAAUGCUACAACAACAGGAGAAAAAAACUAAACC ...--.....(((((.((((((....((((((...((((((....)))))))))))).((((((((......)))))))).)))))))))))............................ ( -28.10) >DroPer_CAF1 160703 102 - 1 GUCUAGGGCCACAUUUGAUGUUCGUUUUUGCAACUAUUUGCAAAUGCAAAUUGCAAACAUGCAACAGGGACAUGUUGCGUAAAUGUCAAUGUUACAACAA----------------AG-- ..........(((((.((((((....((((((...((((((....)))))))))))).((((((((......)))))))).)))))))))))........----------------..-- ( -25.30) >consensus GUC__AAGCCGCAUUUGAUGUUCGUUUUGGCAACUAUUUGCAAAUGCAAAUUGCAAACAUGCAACAGGGACAUGUUGCGUAAAUGUCAAUGUUACAACAA________________AACC ..............((((((((.((((..(((...((((((....)))))))))))))((((((((......)))))))).))))))))............................... (-22.70 = -22.70 + -0.00)

| Location | 20,590,868 – 20,590,965 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -20.76 |
| Energy contribution | -20.52 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
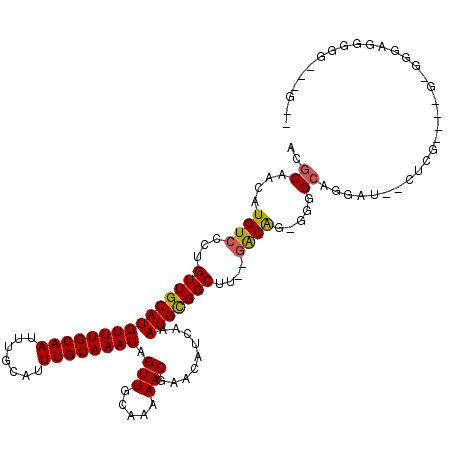
>2L_DroMel_CAF1 20590868 97 + 22407834 ACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCCAAAACGAACAUCAAAUGCGGCUU--GACAGAGGGGCGGGAU--CUCG----A-------------G-- .(((..(.((((...(((((((((((((((.......)))))))).(((.....))).........)))))))..--))))..)..)))....--....----.-------------.-- ( -27.50) >DroPse_CAF1 154600 113 + 1 ACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCAAAAACGAACAUCAAAUGUGGCCCUAGACGG-GGGGCAUAGUGGGUGG----GGGGUAGGGGG--AGUC ((.(..(.(..((((.((((((((((((((.......)))))))))..))))).......((((...((((.((((.....)-))).))))...)))))----)))..).)..)--.)). ( -39.00) >DroEre_CAF1 95951 103 + 1 ACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCCAAAACGAACAUCAAAUGCGGCUU--CGCAG-GCGGCAGGAG--CGCG----G-GGGAGGGG------- ......(...(((((((.((.(((((((((.......)))))))))((((((.....(.....)...((((....--)))))-)))))....)--))))----)-)))...).------- ( -35.10) >DroYak_CAF1 99663 109 + 1 ACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCCAAAACGAACAUCAAAUGCGGCUU--GACAGAGAGGCAGGAU--CGCA----G-GGGAGGGGGCUAG-- ..((..(...((((((((((.(.(((((((.(((((((((......(((.....)))......))))))))).))--).)))).).)))....--.)))----)-)))..)..))...-- ( -34.90) >DroAna_CAF1 99062 112 + 1 ACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCAAAAACGAACAUCAAAUGCGGCUU--GACAAGUCAGGGGGUU--CACCAAAAAGGUGACGGGC--GG-- .(((......((((((((((((.((((((((((((....))))...))))))))............))))))).(--((....)))))))).(--((((.....)))))...))--).-- ( -35.42) >DroPer_CAF1 160725 112 + 1 ACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCAAAAACGAACAUCAAAUGUGGCCCUAGACGG-GGGGCAUAGUGGGUGG----GGGGUAGGGGG--UG-C .(((..(.(..((((.((((((((((((((.......)))))))))..))))).......((((...((((.((((.....)-))).))))...)))))----)))..).)..)--))-. ( -41.90) >consensus ACGCAACAUGUCCCUGUUGCAUGUUUGCAAUUUGCAUUUGCAAAUAGUUGCAAAAACGAACAUCAAAUGCGGCUU__GACAG_GGGGCAGGAU__CUCG____G_GGGAGGGGG___G__ ..((....((((...(((((((((((((((.......)))))))).(((.....))).........)))))))....)))).....))................................ (-20.76 = -20.52 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:07 2006