
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,582,829 – 20,582,921 |
| Length | 92 |
| Max. P | 0.893866 |

| Location | 20,582,829 – 20,582,921 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -10.07 |
| Energy contribution | -10.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
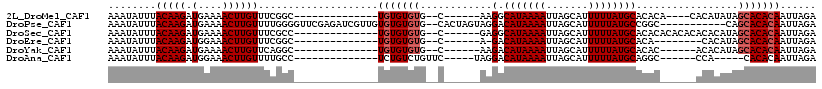
>2L_DroMel_CAF1 20582829 92 + 22407834 AAAUAUUUACAAGAUGAAAACUUGUUUCGGC--------------UGUGUGUG--C------AAGGCAUAAAAUUAGCAUUUUUAUGCACACA----CACAUAUAGCACACAAUUAGA ........(((((.(....))))))....((--------------((((((((--.------...((((((((.......)))))))).....----))))))))))........... ( -25.20) >DroPse_CAF1 145260 105 + 1 AAAUAUUUACAAGAUGAAAACUUGUUUUGGGGUUCGAGAUCGUUGUGUGUGUG--CACUAGUAGGACAUAAAAUUAGCAUUUUUAUGCCGGC-----------CAGCACACAAUUAGA .......(((((((((((..((......))..)))...))).)))))((((((--(....((.((.(((((((.......))))))))).))-----------..)))))))...... ( -24.80) >DroSec_CAF1 97515 96 + 1 AAAUAUUUACAAGAUGAAAACUUGUUUCGCC--------------UGUGUGUG--C------GAGGCAUAAAAUUAGCAUUUUUAUGCACACACACACACACAUAGCACACAAUUAGA ........(((((.(....))))))...((.--------------((((((((--.------(.(((((((((.......)))))))).).).))))))))....))........... ( -24.50) >DroEre_CAF1 88515 87 + 1 AAAUAUUUACAAGAUGGAAACUUGUUUCGGC--------------UGUGUGUG--C------A-GACAUAAAAUUAGCAUUUUUAUGCACA--------CACAUAGCACACAAUUAGA ........(((((.(....))))))....((--------------((((((((--.------.-(.(((((((.......)))))))).))--------))))))))........... ( -23.00) >DroYak_CAF1 91714 90 + 1 AAAUAUUUACAAGAUGAAAACUUGUUCAGGC--------------UGUGUGUG--C------AAGACAUAAAAUUAGCAUUUUUAUGCACAC------ACACAUAGCACACAAUUAGA ........(((((.(....))))))....((--------------((((((((--.------..(.(((((((.......))))))))...)------)))))))))........... ( -23.10) >DroAna_CAF1 90831 88 + 1 AAAUAUUUACAAGAUGGAAACUUGUUUUGCC--------------UCUGUCUGUUC-----UAGGACAUAAAAUUAGCAUUUUUAUGCAGGC------CCA-----CACACAAUUAGA ........(((((.(....))))))...(((--------------(.(((((....-----..)))))........((((....))))))))------...-----............ ( -15.80) >consensus AAAUAUUUACAAGAUGAAAACUUGUUUCGGC______________UGUGUGUG__C______AAGACAUAAAAUUAGCAUUUUUAUGCACAC______ACACAUAGCACACAAUUAGA ........(((((.......)))))....................(((((((............(.(((((((.......)))))))).................)))))))...... (-10.07 = -10.57 + 0.50)
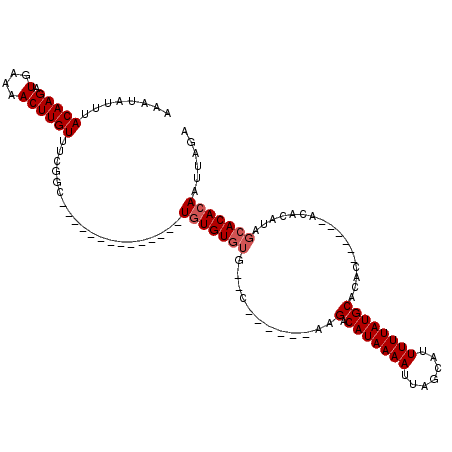
| Location | 20,582,829 – 20,582,921 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -10.93 |
| Energy contribution | -10.88 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
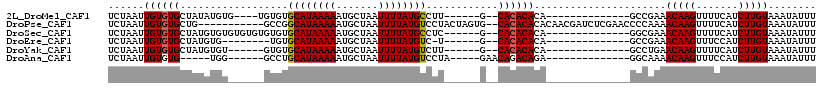
>2L_DroMel_CAF1 20582829 92 - 22407834 UCUAAUUGUGUGCUAUAUGUG----UGUGUGCAUAAAAAUGCUAAUUUUAUGCCUU------G--CACACACA--------------GCCGAAACAAGUUUUCAUCUUGUAAAUAUUU ........((.(((....(((----(((((((((((((.......))))))))...------)--))))))))--------------))))..(((((.......)))))........ ( -22.90) >DroPse_CAF1 145260 105 - 1 UCUAAUUGUGUGCUG-----------GCCGGCAUAAAAAUGCUAAUUUUAUGUCCUACUAGUG--CACACACACAACGAUCUCGAACCCCAAAACAAGUUUUCAUCUUGUAAAUAUUU ......((((..(((-----------(..(((((((((.......))))))).))..))))..--)))).......((....)).........(((((.......)))))........ ( -23.90) >DroSec_CAF1 97515 96 - 1 UCUAAUUGUGUGCUAUGUGUGUGUGUGUGUGCAUAAAAAUGCUAAUUUUAUGCCUC------G--CACACACA--------------GGCGAAACAAGUUUUCAUCUUGUAAAUAUUU ...............(((.((((((((((.((((((((.......))))))))..)------)--))))))))--------------.)))..(((((.......)))))........ ( -31.00) >DroEre_CAF1 88515 87 - 1 UCUAAUUGUGUGCUAUGUG--------UGUGCAUAAAAAUGCUAAUUUUAUGUC-U------G--CACACACA--------------GCCGAAACAAGUUUCCAUCUUGUAAAUAUUU ........((.(((.((((--------(((((((((((.......)))))))).-.------)--)))))).)--------------))))..(((((.......)))))........ ( -22.10) >DroYak_CAF1 91714 90 - 1 UCUAAUUGUGUGCUAUGUGU------GUGUGCAUAAAAAUGCUAAUUUUAUGUCUU------G--CACACACA--------------GCCUGAACAAGUUUUCAUCUUGUAAAUAUUU ...........(((.(((((------(((.((((((((.......))))))))..)------)--)))))).)--------------))....(((((.......)))))........ ( -22.20) >DroAna_CAF1 90831 88 - 1 UCUAAUUGUGUG-----UGG------GCCUGCAUAAAAAUGCUAAUUUUAUGUCCUA-----GAACAGACAGA--------------GGCAAAACAAGUUUCCAUCUUGUAAAUAUUU ............-----...------((((((((....))))........((((...-----.....)))).)--------------)))...(((((.......)))))........ ( -15.30) >consensus UCUAAUUGUGUGCUAUGUGU______GUGUGCAUAAAAAUGCUAAUUUUAUGUCUU______G__CACACACA______________GCCGAAACAAGUUUUCAUCUUGUAAAUAUUU ......((((((..................((((((((.......))))))))............))))))......................(((((.......)))))........ (-10.93 = -10.88 + -0.05)
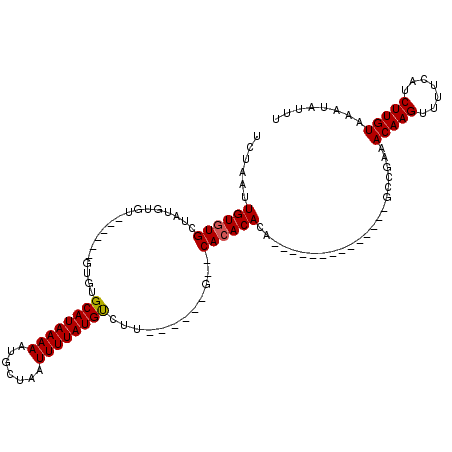
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:36:03 2006