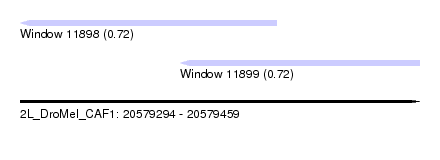
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,579,294 – 20,579,459 |
| Length | 165 |
| Max. P | 0.720144 |

| Location | 20,579,294 – 20,579,400 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.82 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20579294 106 - 22407834 GGCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCAUGUCCGUGUCGGCACGCCCAUGGGCCCCACUCCU-CG-CCCAC---------GUCCCCAUC---AUCGUCAUCCGUGCCC (((((((((((...........))))))).......((.(((.(.(((..((.(((....(((((.........-.)-)))))---------)).))...)---)).).))).)))))). ( -33.80) >DroPse_CAF1 141180 101 - 1 GGCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCAUGUCCAUGUCGGCACGCCCU---GCCACGUAGCA-UG-CCCCC-CCGAA---CUCCCC----------CCCGCUGUACCC (((((((((((...........)))))))).((((..((((((..((((.((((.....)---))))))).)))-))-)....-..)))---).....----------...)))...... ( -33.20) >DroSec_CAF1 93913 106 - 1 GGCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCAUGUCCGUGUCGGCACGCCCAAAAGCCCCACUCCU-CG-CCCAC---------GUCGCCAUC---AUCGCCAUCCGUGCCC (((((((((((...........))))))).......((.(((.(.(((..((((((......((........))-..-....)---------)).)))..)---)).).))).)))))). ( -28.50) >DroEre_CAF1 85214 92 - 1 GGCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCAUGUCCGUGUCGGCACGCCCAUAAGCCCCACUGC--CG-----CC---------------------UCUUCAUCCGUGCCC (((((((((((...........))))))).......((.(((...(.(.(((((.((......)).....)))--))-----.)---------------------.)..))).)))))). ( -26.90) >DroYak_CAF1 88362 115 - 1 GGCAGCCACAGGACCAUAAAUACUGCGGCUUGUUCUCGCAUGUCCGUGUCGGCACGCCCAUAAGCCCCACUCCU-CG-CCCACCCCUUCCUCAUCCCCAUC---AUCGUCAUCCGUGCCC (((((((.(((...........))).)))).......(((((..((((....))))..)))..)).........-.)-)).....................---................ ( -21.70) >DroAna_CAF1 86743 111 - 1 GCCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCAUGUCCGUGUCGGCACGCCCAACGUCCUACGUCCUACGUCCUAC---------GCCCCCAUACGCCCAGGCAUCCAUGCCA ((.((((((((...........)))))))).))....(((((...((((.((.(((....(((.....)))....))))).))---------))........((....))...))))).. ( -33.40) >consensus GGCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCAUGUCCGUGUCGGCACGCCCAUAAGCCCCACUCCU_CG_CCCAC_________GUCCCCAUC___AUCGUCAUCCGUGCCC (((((((((((...........)))))))).((.((.((((....)))).)).))))).............................................................. (-20.62 = -20.82 + 0.20)

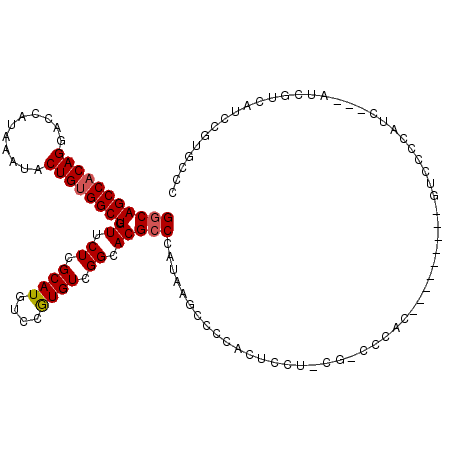
| Location | 20,579,360 – 20,579,459 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -25.22 |
| Energy contribution | -26.18 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20579360 99 - 22407834 AGGAGCA---GCCGGAGCAGGAGC------AGGAGCAGGAGGGGUGCACAUUACUCGUCGCUGUCCCCGGCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCA .((((((---(((((.((....))------.(((.(((((.(((((.....))))).)).)))))))))))((((((((...........)))))))))))))).... ( -44.40) >DroSec_CAF1 93979 90 - 1 AGGAG------------CAGGAGC------AGGAGCAGGCGGGGUGCACAUUACUCGUCGCUGUCCCCGGCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCA .....------------.....((------..((((((.(((((.((((.......)).))...))))).)((((((((...........)))))))))))))..)). ( -37.40) >DroSim_CAF1 94676 102 - 1 AGGAGCAAGAGCAGGAGCAGGAGC------AGGAGCAGGUGGGGUGCACAUUACUCGUCGCUGUCCCCGGCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCA ....((.((((((...((.((..(------((..((..((((........))))..))..)))..))..))((((((((...........)))))))))))))).)). ( -37.30) >DroEre_CAF1 85266 88 - 1 ------------GGGAGCAGGAGC------GGGAGCAGG--GGGCGCACAUUACUCGUCGCUGUCCCCGGCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCA ------------..........((------(((((((((--((((((((.......)).)).))))))...((((((((...........))))))))))))))))). ( -41.70) >DroYak_CAF1 88437 96 - 1 U-----------GGGAGAAGGAGCAGCAAAGGGGGCAGGU-GGAUGCACAUUACUCGUCGCUGUCCCCGGCAGCCACAGGACCAUAAAUACUGCGGCUUGUUCUCGCA .-----------(.(((((.((((.(((..((((((((..-.((((.........)))).))))))))((...((...)).))........))).)))).))))).). ( -37.20) >consensus AGGAG_______GGGAGCAGGAGC______AGGAGCAGGUGGGGUGCACAUUACUCGUCGCUGUCCCCGGCAGCCACAGGACCAUAAAUACUGUGGCUUGUUCUCGCA ...............................(((((((.(((((.((((.......)).))...))))).)((((((((...........)))))))))))))).... (-25.22 = -26.18 + 0.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:58 2006