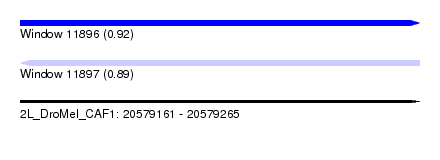
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,579,161 – 20,579,265 |
| Length | 104 |
| Max. P | 0.921032 |

| Location | 20,579,161 – 20,579,265 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.35 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20579161 104 + 22407834 UGCUUCAUUGGCUUGUGCAUUAUUUGUGCAACCUGGGUUGCAUCGUAAAAAAUGCAGAAGCAGCAGGCUGCCACUGCAGCUGGA-----------UCCUUUUGGCCGAGCACACG----- (((((.....((((.((((((.((((((((((....)))))))...))).)))))).)))).((((.......)))).((..((-----------....))..)).)))))....----- ( -37.90) >DroPse_CAF1 141070 98 + 1 UGCUUCAUUGGCUUGUGCAUUAUUUGUGCAACCUGGGUUGCAUCGUAAAAAAUGCACGAGCAGC-GCCUGAAAGUGCAGCUGCU-----------UCCUC--GCCCAUCCAC-------- .........(((..(((((((.((((((((((....)))))))...))).)))))))(((((((-((.(....).)).))))))-----------)....--))).......-------- ( -35.60) >DroEre_CAF1 85069 109 + 1 UGCUUCAUUGGCUUGUGCAUUAUUUGUGCAACCUGGGUUGCAUCGUAAAAAAUGCAGGAGCAGCAGGCUGCCGCUGCAGCUGCC-----------UCCUUUUGGCGGAGCGCACGAGCGG ..........((((((((.((((..(((((((....))))))).))))....(((((..((((....))))..)))))(((.((-----------.((....)).))))))))))))).. ( -47.30) >DroYak_CAF1 88237 104 + 1 UGCUUCAUUGGCUUGUGCAUUAUUUGUGCAACCUGGGUUGCAUCGUAAAAAAUGCAGGAGCAGCAGGCUGCCGCUGCAGCUGGA-----------UCCUUUUGGCUGAGCACACG----- .((((.....((((.((((((.((((((((((....)))))))...))).)))))).))))...))))....(((.((((..((-----------....))..))))))).....----- ( -40.50) >DroAna_CAF1 86585 103 + 1 UGCUUCAUUGGCUUGUGCAUUAUUUGUGCAACCUGGGUUGCAUCGUAAAAAAUGCAGGAGCAGC-GCCUGCAACGGCAGCUGCUACUCCUUGCUUUCCUUUCAG---------------- .((.......((((.((((((.((((((((((....)))))))...))).)))))).))))(((-(.((((....)))).)))).......))...........---------------- ( -34.70) >DroPer_CAF1 147289 98 + 1 UGCUUCAUUGGCUUGUGCAUUAUUUGUGCAACCUGGGUUGCAUCGUAAAAAAUGCACGAGCAGC-GCCUGAAAGUGCAGCUGCU-----------UCCUC--GCCCAUCCAC-------- .........(((..(((((((.((((((((((....)))))))...))).)))))))(((((((-((.(....).)).))))))-----------)....--))).......-------- ( -35.60) >consensus UGCUUCAUUGGCUUGUGCAUUAUUUGUGCAACCUGGGUUGCAUCGUAAAAAAUGCAGGAGCAGC_GCCUGCAACUGCAGCUGCU___________UCCUUUUGGCCGAGCAC________ ..........((((.((((((.((((((((((....)))))))...))).)))))).)))).((.(.((((....))))).))..................................... (-25.82 = -26.35 + 0.53)


| Location | 20,579,161 – 20,579,265 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20579161 104 - 22407834 -----CGUGUGCUCGGCCAAAAGGA-----------UCCAGCUGCAGUGGCAGCCUGCUGCUUCUGCAUUUUUUACGAUGCAACCCAGGUUGCACAAAUAAUGCACAAGCCAAUGAAGCA -----..((((((((.((....)).-----------......(((((.(((((....))))).))))).......)))((((((....))))))........))))).((.......)). ( -33.60) >DroPse_CAF1 141070 98 - 1 --------GUGGAUGGGC--GAGGA-----------AGCAGCUGCACUUUCAGGC-GCUGCUCGUGCAUUUUUUACGAUGCAACCCAGGUUGCACAAAUAAUGCACAAGCCAAUGAAGCA --------((.....(((--...((-----------.(((((.((........))-)))))))(((((((.(((....((((((....)))))).))).)))))))..)))......)). ( -35.20) >DroEre_CAF1 85069 109 - 1 CCGCUCGUGCGCUCCGCCAAAAGGA-----------GGCAGCUGCAGCGGCAGCCUGCUGCUCCUGCAUUUUUUACGAUGCAACCCAGGUUGCACAAAUAAUGCACAAGCCAAUGAAGCA ..(((.(((((((((.......)))-----------)((((..(((((((....)))))))..)))).........(.((((((....)))))))......))))).))).......... ( -42.50) >DroYak_CAF1 88237 104 - 1 -----CGUGUGCUCAGCCAAAAGGA-----------UCCAGCUGCAGCGGCAGCCUGCUGCUCCUGCAUUUUUUACGAUGCAACCCAGGUUGCACAAAUAAUGCACAAGCCAAUGAAGCA -----..(((((.......((((((-----------(.(((..(((((((....)))))))..))).)))))))..(.((((((....))))))).......))))).((.......)). ( -33.90) >DroAna_CAF1 86585 103 - 1 ----------------CUGAAAGGAAAGCAAGGAGUAGCAGCUGCCGUUGCAGGC-GCUGCUCCUGCAUUUUUUACGAUGCAACCCAGGUUGCACAAAUAAUGCACAAGCCAAUGAAGCA ----------------((....))...((.(((((((((..((((....))))..-)))))))))(((((.(((....((((((....)))))).))).))))).............)). ( -36.60) >DroPer_CAF1 147289 98 - 1 --------GUGGAUGGGC--GAGGA-----------AGCAGCUGCACUUUCAGGC-GCUGCUCGUGCAUUUUUUACGAUGCAACCCAGGUUGCACAAAUAAUGCACAAGCCAAUGAAGCA --------((.....(((--...((-----------.(((((.((........))-)))))))(((((((.(((....((((((....)))))).))).)))))))..)))......)). ( -35.20) >consensus ________GUGCUCGGCCAAAAGGA___________AGCAGCUGCAGUGGCAGCC_GCUGCUCCUGCAUUUUUUACGAUGCAACCCAGGUUGCACAAAUAAUGCACAAGCCAAUGAAGCA ........................................(((((((..((((....))))..)))).........(.((((((....))))))).....................))). (-22.26 = -22.32 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:57 2006