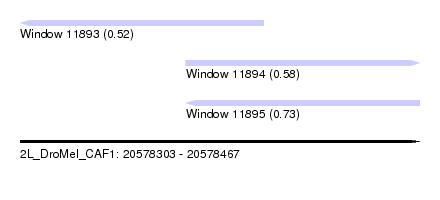
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,578,303 – 20,578,467 |
| Length | 164 |
| Max. P | 0.729397 |

| Location | 20,578,303 – 20,578,403 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -15.09 |
| Energy contribution | -16.40 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20578303 100 - 22407834 ---CGGCGGCAG-----GACAGCAGCAACAUGUUGCUGACAGUUGUUCAAUUCGCAU-CAGA-AAGUUGGCUCAACUGUUGUGGCCUUUGUGCCAUCAUUU-UUGCAUUAA ---.(((.(((.-----..(((((((.....)))))))(((((((.((((((..(..-..).-.))))))..)))))))))).)))...((((.(......-).))))... ( -33.20) >DroSec_CAF1 92929 86 - 1 ----------------------CAGCAACAUGUUGCUGACAGUUGUUCAAUUCGCAU-CUGA-AAGUUGGCUCAACUGUUGUGGCCUUUGUGCCAUCAUUU-UUGCAUUAA ----------------------((((((....))))))(((((((.((((((..(..-..).-.))))))..))))))).(((((......))))).....-......... ( -24.90) >DroSim_CAF1 93616 86 - 1 ----------------------CAGCAACAUGUUGCUGACAGUUGUUCAAUUCGCAU-CUGA-AAGUUGGUUCAACUUUUGUGGCCUUUGUGCCAUCAUUU-UUGCAUUAA ----------------------((((((....))))))...............(((.-..((-((((((...))))))))(((((......))))).....-.)))..... ( -21.30) >DroEre_CAF1 84340 102 - 1 CGGCGGCGGCCG-----UACAGCAGCAACAUGUUGCUGACAGUUGUUCGAUUCGCAU-CAGAAAAGUUGGCUCCGCUGUUGUGGCCUUUGUGCCAUC--UU-UUGCAUUAA ((((((.(((((-----(.(((((((.....)))))))))..........(((....-..))).....))))))))))..(((((......))))).--..-......... ( -34.70) >DroYak_CAF1 87350 100 - 1 ----GGCGGCAG-----GACAGCAGCAACAUGUUGCUGACAGUUGUUCAAUUCGCAU-CAGAAAAGUUGCCUUAACUGUUGUGGCCUUUGUGCCAUCAUUU-UUGCAUUAA ----(((.(((.-----..(((((((.....)))))))(((((((..((((((....-..))...))))...)))))))))).)))...((((.(......-).))))... ( -28.90) >DroPer_CAF1 144668 103 - 1 ---CGGUGGGUGGUUGCCACAACAGCAACAUGUUGCUGACAGUUGUUCAAUUCGCAUACAGAAAAAAU-----AAAAGAGAAGUCCUUUUGGCCAACAUUUUCUGCAUUAA ---..((((((((...))))..((((((....)))))).............))))...(((((((..(-----(((((.(....))))))).......)))))))...... ( -21.50) >consensus ____GGCGGC_G______ACAGCAGCAACAUGUUGCUGACAGUUGUUCAAUUCGCAU_CAGA_AAGUUGGCUCAACUGUUGUGGCCUUUGUGCCAUCAUUU_UUGCAUUAA ......................((((((....))))))(((((((..(((((..(.....)...)))))...))))))).(((((......)))))............... (-15.09 = -16.40 + 1.31)

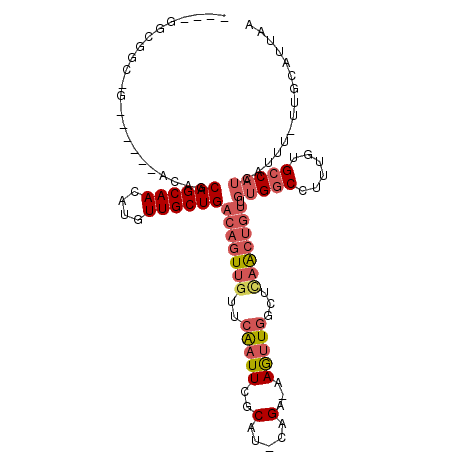
| Location | 20,578,371 – 20,578,467 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 71.82 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20578371 96 + 22407834 GUCAGCAACAUGUUGCUGCUGUC-----CUGCCGCCGCCGCCUUUCCCCACCCGGAAAAC-CCGGAAAACU-ACCGCUUUUCCGCCUUGGAGGCC-GGCGCCUG (.((((((....)))))))....-----..(.(((((..((((....(((..(((((((.-.(((......-.))).)))))))...))))))))-)))).).. ( -32.30) >DroPse_CAF1 138437 94 + 1 GUCAGCAACAUGUUGCUGUUGUGGCAAUCACCCACCGUGGCACCAC---ACCC-------ACCCCCCGCCUGUCUGCCUGCCCGCCUUCAAGGCCUGGCAUUUG ..((((((....))))))....((((..((......((((......---..))-------))........))..))))((((.(((.....)))..)))).... ( -26.04) >DroSec_CAF1 92997 82 + 1 GUCAGCAACAUGUUGCUG-------------------CCGCCUUUCUCCACCCGGAAAAC-CCUGAAAACU-ACCGCUUUUCCGCCUUGGAGGCC-GGCGCUUG ..((((((....))))))-------------------.((((..((((((..(((((((.-..((......-..)).)))))))...))))))..-)))).... ( -27.40) >DroSim_CAF1 93684 82 + 1 GUCAGCAACAUGUUGCUG-------------------CCGCCUUUCUCCACCCGGAAAAC-CCGGAAAACU-ACCGCUUUUCCGCCUUGGAGGCC-GGCGCUUG ..((((((....))))))-------------------.((((..((((((..(((((((.-.(((......-.))).)))))))...))))))..-)))).... ( -31.80) >DroYak_CAF1 87419 93 + 1 GUCAGCAACAUGUUGCUGCUGUC-----CUGCCGCC---GACUUUCCCCACCCGGAAAAC-CCGGAAAACU-ACCGCCUUUCCGCCUUGGAGGCC-GGCGUUUG (.((((((....)))))))....-----....((((---(.(((((......((((((..-.(((......-.)))..))))))....))))).)-)))).... ( -29.10) >consensus GUCAGCAACAUGUUGCUG_UGU______C__CC_CC_CCGCCUUUCCCCACCCGGAAAAC_CCGGAAAACU_ACCGCUUUUCCGCCUUGGAGGCC_GGCGCUUG ..((((((....)))))).................................((((......)))).........((((.....(((.....)))..)))).... (-16.88 = -17.32 + 0.44)


| Location | 20,578,371 – 20,578,467 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 71.82 |
| Mean single sequence MFE | -36.56 |
| Consensus MFE | -18.64 |
| Energy contribution | -21.56 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20578371 96 - 22407834 CAGGCGCC-GGCCUCCAAGGCGGAAAAGCGGU-AGUUUUCCGG-GUUUUCCGGGUGGGGAAAGGCGGCGGCGGCAG-----GACAGCAGCAACAUGUUGCUGAC ..(.((((-(.((((((...(((((((.(((.-......)).)-.)))))))..)))))...).))))).).....-----..(((((((.....))))))).. ( -41.40) >DroPse_CAF1 138437 94 - 1 CAAAUGCCAGGCCUUGAAGGCGGGCAGGCAGACAGGCGGGGGGU-------GGGU---GUGGUGCCACGGUGGGUGAUUGCCACAACAGCAACAUGUUGCUGAC ....((((..(((.....))).))))((((..((.((.......-------..))---.)).))))...((((.......))))..((((((....)))))).. ( -33.60) >DroSec_CAF1 92997 82 - 1 CAAGCGCC-GGCCUCCAAGGCGGAAAAGCGGU-AGUUUUCAGG-GUUUUCCGGGUGGAGAAAGGCGG-------------------CAGCAACAUGUUGCUGAC ....((((-...(((((...(((((((.(...-.........)-.)))))))..)))))...)))).-------------------((((((....)))))).. ( -32.20) >DroSim_CAF1 93684 82 - 1 CAAGCGCC-GGCCUCCAAGGCGGAAAAGCGGU-AGUUUUCCGG-GUUUUCCGGGUGGAGAAAGGCGG-------------------CAGCAACAUGUUGCUGAC ....((((-...(((((...(((((((.(((.-......)).)-.)))))))..)))))...)))).-------------------((((((....)))))).. ( -34.80) >DroYak_CAF1 87419 93 - 1 CAAACGCC-GGCCUCCAAGGCGGAAAGGCGGU-AGUUUUCCGG-GUUUUCCGGGUGGGGAAAGUC---GGCGGCAG-----GACAGCAGCAACAUGUUGCUGAC ....((((-((((((((...(((((((.(((.-......)).)-.)))))))..)))))...)))---))))....-----..(((((((.....))))))).. ( -40.80) >consensus CAAGCGCC_GGCCUCCAAGGCGGAAAAGCGGU_AGUUUUCCGG_GUUUUCCGGGUGGAGAAAGGCGG_GG_GG__G______ACA_CAGCAACAUGUUGCUGAC ....((((....(((((...(((((((.(((........)))...)))))))..)))))...))))....................((((((....)))))).. (-18.64 = -21.56 + 2.92)

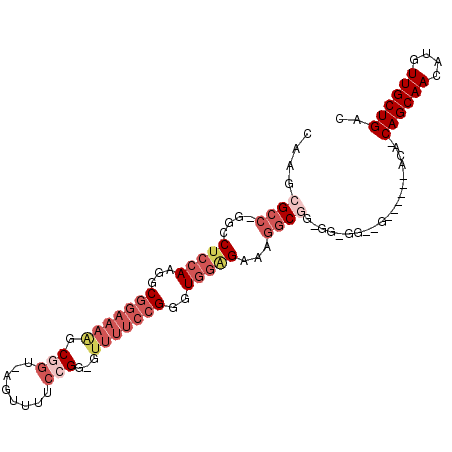
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:55 2006