
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,577,580 – 20,577,684 |
| Length | 104 |
| Max. P | 0.887535 |

| Location | 20,577,580 – 20,577,684 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.05 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20577580 104 + 22407834 GGACAACAUGAUGGAGCUCAUCAACAUCAGUCGGCGGGAUGUAUGUGCACGUAUACAUGCCCCUAUGAUGCCCAAGAU-------GGGGGCCUGGA--------AAUGCCGAUGGUCUU ((((....(((((...........)))))(((((((...((((((....))))))((.((((((((..........))-------)))))).))..--------..))))))).)))). ( -35.90) >DroSec_CAF1 92207 101 + 1 GGACAAAAUGAUGGAGCUCAUCAACAUCAGUCGGCGGGAUGUAUGUGCACGUAUACAUGCCCCCA---UGCCCAAGAU-------GGGCGCCUGGA--------ACUGCCGAUGGUCUU ((((....(((((...........)))))((((((((((((((((......))))))).)))(((---(((((.....-------)))))..))).--------...)))))).)))). ( -38.70) >DroSim_CAF1 92896 101 + 1 GGACAACAUGAUGGAGCUCAUCAACAUCAGUCGGCGGGAUGUAUGUGCACGUAUACAUGCCCCCA---UGCCCAAGAU-------GGGCGCCUGGA--------ACUGCCGAUGGUCUU ((((....(((((...........)))))((((((((((((((((......))))))).)))(((---(((((.....-------)))))..))).--------...)))))).)))). ( -38.70) >DroEre_CAF1 83721 93 + 1 GCACAACAUGAUGGAGCUCAUCAGCAUCAGUCGGCGGGAUGUAUGUGCACGUAUACACGCCC------GG-CCGGGAU-------GG-GGCC---A--------ACUGCCGAUGGUGCC ........(((((.....)))))((((((.((((((((.((((((......))))))..)))------((-((.....-------..-))))---.--------...))))))))))). ( -37.90) >DroYak_CAF1 86644 95 + 1 GGACAACAUGAUGGAGCUCAUCAACAUCAGUCGGCGGGAUAUAUGUGCACGUAUACAUGCCA------UGCCCAAGAU-------GGGGGCC---A--------ACUGCCGAUGGUCUU ((((....(((((...........)))))((((((((..((((((....))))))...(((.------..(((.....-------)))))).---.--------.)))))))).)))). ( -31.80) >DroAna_CAF1 84819 105 + 1 AGACAACAUGAUGGAGCUCAUCAACAUCAGAUGAGGAGACAGAA-----------AAUGGU---AGGGGGGUCGAAAAGGAGGUAGAGGUCCUGGAGCAUCAGGAAUGCCAAUGGUCUU ((((..(((..((...((((((.......))))))....))...-----------.)))..---.................((((....(((((......))))).))))....)))). ( -24.30) >consensus GGACAACAUGAUGGAGCUCAUCAACAUCAGUCGGCGGGAUGUAUGUGCACGUAUACAUGCCC__A___UGCCCAAGAU_______GGGGGCCUGGA________ACUGCCGAUGGUCUU ((((....(((((...........)))))((((((((.(((((((......))))))).)).........(((............)))...................)))))).)))). (-20.86 = -21.70 + 0.84)

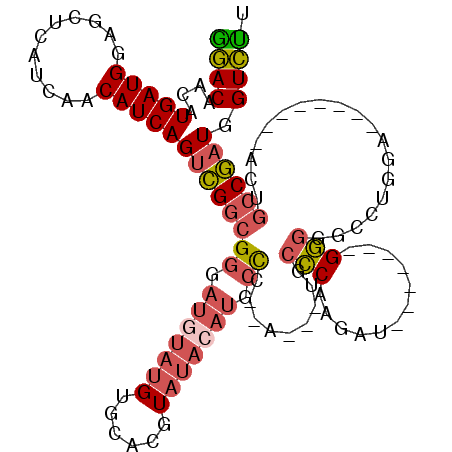
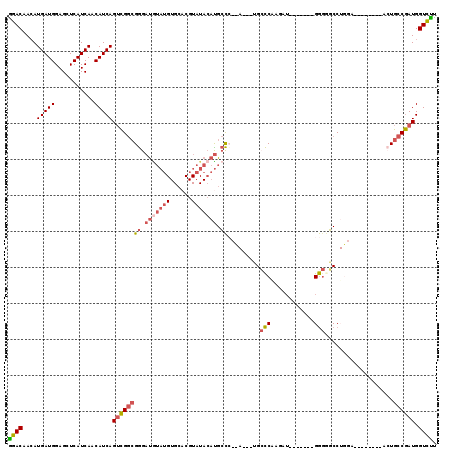
| Location | 20,577,580 – 20,577,684 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.05 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20577580 104 - 22407834 AAGACCAUCGGCAUU--------UCCAGGCCCCC-------AUCUUGGGCAUCAUAGGGGCAUGUAUACGUGCACAUACAUCCCGCCGACUGAUGUUGAUGAGCUCCAUCAUGUUGUCC ..(((((((((((((--------....(((.(((-------.....)))........(((.((((((........))))))))))))....))))))))))(((........)))))). ( -32.70) >DroSec_CAF1 92207 101 - 1 AAGACCAUCGGCAGU--------UCCAGGCGCCC-------AUCUUGGGCA---UGGGGGCAUGUAUACGUGCACAUACAUCCCGCCGACUGAUGUUGAUGAGCUCCAUCAUUUUGUCC ..((((((((((...--------.(((...((((-------.....)))).---)))(((.((((((........)))))))))))))).))....(((((.....)))))....))). ( -35.50) >DroSim_CAF1 92896 101 - 1 AAGACCAUCGGCAGU--------UCCAGGCGCCC-------AUCUUGGGCA---UGGGGGCAUGUAUACGUGCACAUACAUCCCGCCGACUGAUGUUGAUGAGCUCCAUCAUGUUGUCC ..(((((((((((..--------....(((((((-------.....)))).---...(((.((((((........))))))))))))......))))))))(((........)))))). ( -35.90) >DroEre_CAF1 83721 93 - 1 GGCACCAUCGGCAGU--------U---GGCC-CC-------AUCCCGG-CC------GGGCGUGUAUACGUGCACAUACAUCCCGCCGACUGAUGCUGAUGAGCUCCAUCAUGUUGUGC ((((.(((((((...--------.---((((-..-------.....))-))------(((.((((((........)))))))))))))).)).))))((((.....))))......... ( -34.00) >DroYak_CAF1 86644 95 - 1 AAGACCAUCGGCAGU--------U---GGCCCCC-------AUCUUGGGCA------UGGCAUGUAUACGUGCACAUAUAUCCCGCCGACUGAUGUUGAUGAGCUCCAUCAUGUUGUCC ..(((((((((((((--------(---(((.(((-------.....))).(------(((((((....))))).))).......))))))...))))))))(((........)))))). ( -31.10) >DroAna_CAF1 84819 105 - 1 AAGACCAUUGGCAUUCCUGAUGCUCCAGGACCUCUACCUCCUUUUCGACCCCCCU---ACCAUU-----------UUCUGUCUCCUCAUCUGAUGUUGAUGAGCUCCAUCAUGUUGUCU .(((((((..(((((...((((....((((........))))....(((......---......-----------....)))....)))).)))))..)))(((........))))))) ( -18.29) >consensus AAGACCAUCGGCAGU________UCCAGGCCCCC_______AUCUUGGGCA___U__GGGCAUGUAUACGUGCACAUACAUCCCGCCGACUGAUGUUGAUGAGCUCCAUCAUGUUGUCC ..((((((((((...................(((............)))........(((.((((((........)))))))))))))).))....(((((.....)))))....))). (-17.57 = -17.65 + 0.08)
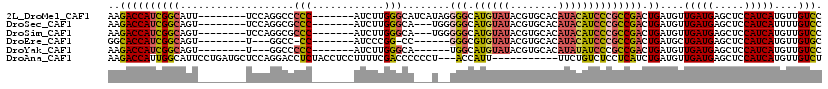
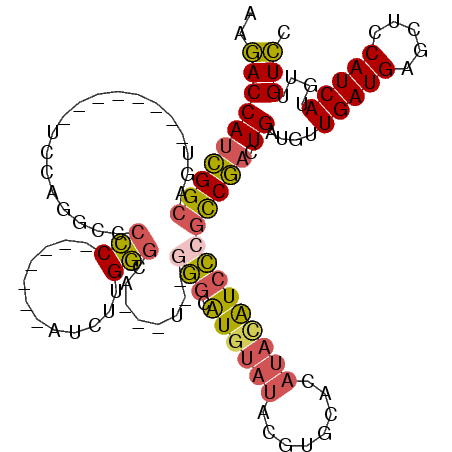
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:52 2006