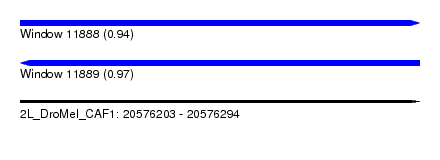
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,576,203 – 20,576,294 |
| Length | 91 |
| Max. P | 0.974939 |

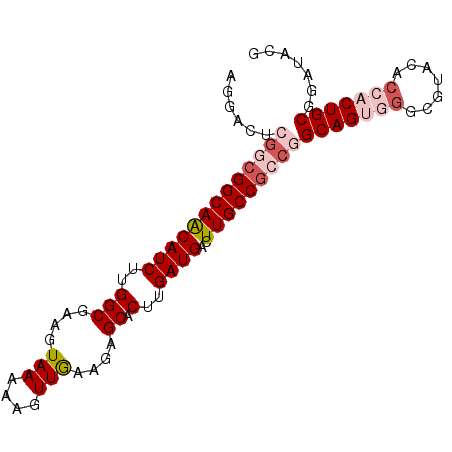
| Location | 20,576,203 – 20,576,294 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -24.07 |
| Energy contribution | -25.51 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20576203 91 + 22407834 CGUAUCCGCAGUGGUGUACGCCCACUGCCGGCGGCAAGUCAUCAAGUGCUCUUCAACUUUUUACUUCGCCAAGAUGUUGCCGCCGAGUACU .((((..(((((((.(....))))))))(((((((((..((((..(.((..................)))..))))))))))))).)))). ( -34.87) >DroSec_CAF1 90841 91 + 1 CGUAUCCGCAGUGGUGUACGCCCACUGCCGGCGGCAAGUCAUCAAGUGCUCUUCAACUUUUUACUUCGCCAAGAUGUUGCCGUCGAGUCCU .......(((((((.(....))))))))(((((((((..((((..(.((..................)))..)))))))))))))...... ( -30.47) >DroSim_CAF1 91534 91 + 1 CGUAUCCGCAGUGGUGUACGCCCACUGCCGGCGGCAAGUCAUCAAGUGCUCUUCAACUUUUUACUUCGCCAAGAUGUUGCCGCCGAGUCCU .......(((((((.(....))))))))(((((((((..((((..(.((..................)))..)))))))))))))...... ( -33.17) >DroEre_CAF1 82469 85 + 1 CGUAUCCGCAGUGGUGUACGCC--CUGCU---GGCAAGUCAUCAAGUGCGCUUCAACUU-UUACUUGGCCAAGAUGCUGCCGCCGAGUCCU .(((((.((((.(((....)))--))))(---(((((((....((((........))))-..)))).)))).))))).............. ( -24.20) >DroYak_CAF1 85288 85 + 1 CGUAUCCGCAGUGGCCCA------CUGCUGGCGGCAAGUCAUCAAGUGCUCUUUAACUUUUUACUUCGCCAAGAUGUUGCCGCCGAGUCCU .......((((((...))------))))(((((((((..((((..(.((..................)))..)))))))))))))...... ( -26.47) >consensus CGUAUCCGCAGUGGUGUACGCCCACUGCCGGCGGCAAGUCAUCAAGUGCUCUUCAACUUUUUACUUCGCCAAGAUGUUGCCGCCGAGUCCU .......(((((((.......)))))))(((((((((..((((..(.((..................)))..)))))))))))))...... (-24.07 = -25.51 + 1.44)

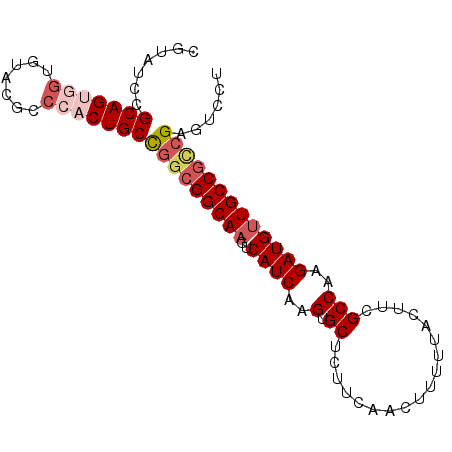
| Location | 20,576,203 – 20,576,294 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -26.92 |
| Energy contribution | -28.80 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20576203 91 - 22407834 AGUACUCGGCGGCAACAUCUUGGCGAAGUAAAAAGUUGAAGAGCACUUGAUGACUUGCCGCCGGCAGUGGGCGUACACCACUGCGGAUACG .(((..(((((((((((((..(((....(((....)))....)).)..))))..)))))))).(((((((.......))))))))..))). ( -39.30) >DroSec_CAF1 90841 91 - 1 AGGACUCGACGGCAACAUCUUGGCGAAGUAAAAAGUUGAAGAGCACUUGAUGACUUGCCGCCGGCAGUGGGCGUACACCACUGCGGAUACG ......((.((((((((((..(((....(((....)))....)).)..))))..)))))).))(((((((.......)))))))....... ( -30.30) >DroSim_CAF1 91534 91 - 1 AGGACUCGGCGGCAACAUCUUGGCGAAGUAAAAAGUUGAAGAGCACUUGAUGACUUGCCGCCGGCAGUGGGCGUACACCACUGCGGAUACG ......(((((((((((((..(((....(((....)))....)).)..))))..)))))))))(((((((.......)))))))....... ( -37.40) >DroEre_CAF1 82469 85 - 1 AGGACUCGGCGGCAGCAUCUUGGCCAAGUAA-AAGUUGAAGCGCACUUGAUGACUUGCC---AGCAG--GGCGUACACCACUGCGGAUACG ........((....))((((((((.((((..-((((........))))....)))))))---)((((--((......)).))))))))... ( -21.40) >DroYak_CAF1 85288 85 - 1 AGGACUCGGCGGCAACAUCUUGGCGAAGUAAAAAGUUAAAGAGCACUUGAUGACUUGCCGCCAGCAG------UGGGCCACUGCGGAUACG .......((((((((((((..(((....(((....)))....)).)..))))..)))))))).((((------((...))))))....... ( -30.60) >consensus AGGACUCGGCGGCAACAUCUUGGCGAAGUAAAAAGUUGAAGAGCACUUGAUGACUUGCCGCCGGCAGUGGGCGUACACCACUGCGGAUACG ......(((((((((((((..(((....(((....)))....)).)..))))..)))))))))(((((((.......)))))))....... (-26.92 = -28.80 + 1.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:48 2006