
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,573,346 – 20,573,509 |
| Length | 163 |
| Max. P | 0.990086 |

| Location | 20,573,346 – 20,573,447 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.23 |
| Mean single sequence MFE | -35.65 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.08 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
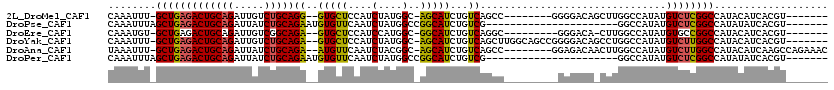
>2L_DroMel_CAF1 20573346 101 + 22407834 CAAAUUU-GCUGAGACUGCAGAUUGUCUGCAGG--GUGCUCCAUCUAUGGC-AGCAUCUGUCAGCC--------GGGGACAGCUUGGCCAUAUGUCUCGGCCAUACAUCACGU------- .......-(((((((((((((.....)))))((--((((((((....))).-)))))))((((((.--------(....).)).)))).....))))))))............------- ( -39.40) >DroPse_CAF1 133102 91 + 1 CAAAUUUAGCUGAGACUGCAGAUUAUCUGCAGAAUGUGUUCAAUCUAUGGCCGGCAUCUGUCG----------------------GGCCAUAUGUCUCGGCCAUAUAUCACGU------- ........(((((((((((((.....)))))).............((((((((((....))).----------------------)))))))..)))))))............------- ( -33.80) >DroEre_CAF1 80054 99 + 1 CAAAUGU-GCUGAGACUGCAGAUUGUCGGCAGA--GUGCUCCAUCCAUGGC-GGCAUCUGUCAGGC---------GGGACA-CUUGGCCAUAUGUGCCGGCCAUACAUCACGU------- ...((((-(.((.(((........)))((((((--.(((((((....))).-)))))))))).(((---------.((.((-(..........))))).)))...)).)))))------- ( -35.40) >DroYak_CAF1 82495 109 + 1 CAAAUUU-GCUGAGACUGCAGAUUGUCUGCAGA--GUGCUCCAUCUAUGGC-AGCAUCUGUCAGCUUGGCAGCCGGGGACAGCCUGGCCAUAUGUCUUGGCCAUACAUCACGU------- .......-(((((.(((((((.....)))))).--((((((((....))).-))))).).))))).((((.((((((.....))))))((.......))))))..........------- ( -40.00) >DroAna_CAF1 80434 108 + 1 UAAAUUU-GCUGAGACUGCAGAUUAUCUGCAGA--AUGUUCAAUCUACGGC-AGCAUCUGUCAGCC--------GGAGACAACUUGGCCAUAUGUCUUGGCCAUACAUCAAGCCAGAAAC .....((-(((((((((((((.....)))))).--........))).))))-))..((((.(....--------(....)....((((((.......))))))........).))))... ( -31.50) >DroPer_CAF1 139011 91 + 1 CAAAUUUAGCUGAGACUGCAGAUUAUCUGCAGAAUGUGUUCAAUCUAUGGCCGGCAUCUGUCG----------------------GGCCAUAUGUCUCGGCCAUAUAUCACGU------- ........(((((((((((((.....)))))).............((((((((((....))).----------------------)))))))..)))))))............------- ( -33.80) >consensus CAAAUUU_GCUGAGACUGCAGAUUAUCUGCAGA__GUGCUCAAUCUAUGGC_AGCAUCUGUCAGCC_________GGGACA_CUUGGCCAUAUGUCUCGGCCAUACAUCACGU_______ ........(((((((((((((.....)))))((..(((((............)))))...))...............................))))))))................... (-20.58 = -20.08 + -0.50)

| Location | 20,573,346 – 20,573,447 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.23 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
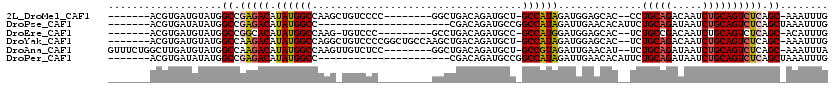
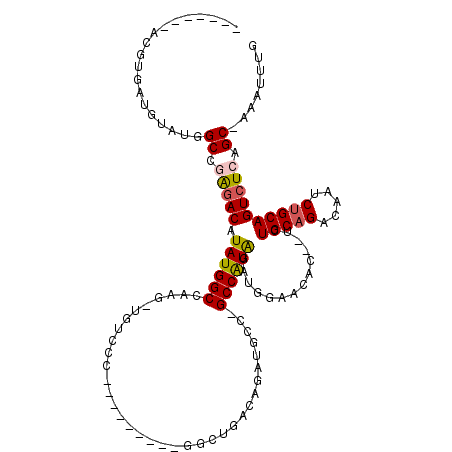
>2L_DroMel_CAF1 20573346 101 - 22407834 -------ACGUGAUGUAUGGCCGAGACAUAUGGCCAAGCUGUCCCC--------GGCUGACAGAUGCU-GCCAUAGAUGGAGCAC--CCUGCAGACAAUCUGCAGUCUCAGC-AAAUUUG -------.((.(((((.((((((.......)))))).)).)))..)--------)(((((....((((-.(((....))))))).--.((((((.....))))))..)))))-....... ( -34.60) >DroPse_CAF1 133102 91 - 1 -------ACGUGAUAUAUGGCCGAGACAUAUGGCC----------------------CGACAGAUGCCGGCCAUAGAUUGAACACAUUCUGCAGAUAAUCUGCAGUCUCAGCUAAAUUUG -------..........((((.(((((.(((((((----------------------.(.(....)).)))))))..............(((((.....)))))))))).))))...... ( -30.20) >DroEre_CAF1 80054 99 - 1 -------ACGUGAUGUAUGGCCGGCACAUAUGGCCAAG-UGUCCC---------GCCUGACAGAUGCC-GCCAUGGAUGGAGCAC--UCUGCCGACAAUCUGCAGUCUCAGC-ACAUUUG -------..(((((.((((((.((((....((..((.(-((...)---------)).)).))..))))-)))))).)).(((...--.((((.(.....).)))).)))..)-))..... ( -28.10) >DroYak_CAF1 82495 109 - 1 -------ACGUGAUGUAUGGCCAAGACAUAUGGCCAGGCUGUCCCCGGCUGCCAAGCUGACAGAUGCU-GCCAUAGAUGGAGCAC--UCUGCAGACAAUCUGCAGUCUCAGC-AAAUUUG -------..........((((((.......))))))(((.((.....)).)))..(((((.((.((((-.(((....))))))))--)((((((.....))))))..)))))-....... ( -39.10) >DroAna_CAF1 80434 108 - 1 GUUUCUGGCUUGAUGUAUGGCCAAGACAUAUGGCCAAGUUGUCUCC--------GGCUGACAGAUGCU-GCCGUAGAUUGAACAU--UCUGCAGAUAAUCUGCAGUCUCAGC-AAAUUUA ((((((((...((((..((((((.......))))))...))))...--------(((((.(((((.((-((...(((........--)))))))...)))))))))))))).-))))... ( -32.60) >DroPer_CAF1 139011 91 - 1 -------ACGUGAUAUAUGGCCGAGACAUAUGGCC----------------------CGACAGAUGCCGGCCAUAGAUUGAACACAUUCUGCAGAUAAUCUGCAGUCUCAGCUAAAUUUG -------..........((((.(((((.(((((((----------------------.(.(....)).)))))))..............(((((.....)))))))))).))))...... ( -30.20) >consensus _______ACGUGAUGUAUGGCCGAGACAUAUGGCCAAG_UGUCCC_________GGCUGACAGAUGCC_GCCAUAGAUGGAACAC__UCUGCAGACAAUCUGCAGUCUCAGC_AAAUUUG ...................((.(((((.((((((...................................))))))..............(((((.....)))))))))).))........ (-18.92 = -19.17 + 0.25)

| Location | 20,573,383 – 20,573,474 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.18 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -14.57 |
| Energy contribution | -15.91 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20573383 91 + 22407834 CCAUCUAUGGCAGCAUCUGUCAGCC--------GGGGACAGCUUGGCCAUAUGUCUCGGCCAUACAUCACGU-------ACACAUAGUAUCU----UUGUGUGUACUGGC--------- (((..((((((.((((..((((((.--------(....).)).))))...))))....))))))......((-------((((((((.....----))))))))))))).--------- ( -31.80) >DroSec_CAF1 88140 89 + 1 CCAUCUAUGGCAGCAUCUGUCAGCC--------GGGGACAGCUUGGCCAUAUGUCUCGGCCAUACAUCACGU-------ACACAUAGUAUCU------GUGUGUGCUGGC--------- (((..((((((.((((..((((((.--------(....).)).))))...))))....))))))......((-------((((((((...))------))))))))))).--------- ( -32.10) >DroSim_CAF1 88815 89 + 1 CCAUCUAUGGCAGCAUCUGUCAGCC--------GGGGACAGCUUGGCCAUAUGUCUCGGCCAUACAUCACGU-------ACACAUAGUAUCU------GUGUGUACUGGC--------- (((..((((((.((((..((((((.--------(....).)).))))...))))....))))))......((-------((((((((...))------))))))))))).--------- ( -32.10) >DroEre_CAF1 80091 93 + 1 CCAUCCAUGGCGGCAUCUGUCAGGC---------GGGACA-CUUGGCCAUAUGUGCCGGCCAUACAUCACGU-------ACAUAUGGUAUGUAUGUACUGCUGUACUGGC--------- (((....)))((((((.(((..(((---------.((...-.)).)))))).))))))((((((((.((.((-------((((((.....)))))))))).)))).))))--------- ( -32.80) >DroYak_CAF1 82532 98 + 1 CCAUCUAUGGCAGCAUCUGUCAGCUUGGCAGCCGGGGACAGCCUGGCCAUAUGUCUUGGCCAUACAUCACGU-------AUAUG-----UGUAUGUACGGGUGUGCUGGC--------- (((....((((((...))))))...)))..(((((.(.((.((((((((.......))))))(((((((((.-------...))-----)).))))).)).))).)))))--------- ( -37.10) >DroAna_CAF1 80471 101 + 1 CAAUCUACGGCAGCAUCUGUCAGCC--------GGAGACAACUUGGCCAUAUGUCUUGGCCAUACAUCAAGCCAGAAACAUACAUAGCACU--------G--GCACUGGCUACGAGGCU ........(((((...)))))((((--------((.((.....((((((.......))))))....))..(((((..............))--------)--)).))))))........ ( -31.44) >consensus CCAUCUAUGGCAGCAUCUGUCAGCC________GGGGACAGCUUGGCCAUAUGUCUCGGCCAUACAUCACGU_______ACACAUAGUAUCU______GGGUGUACUGGC_________ .....((((((.....(((((..((........)).)))))....)))))).......(((((((((.................................))))).))))......... (-14.57 = -15.91 + 1.34)


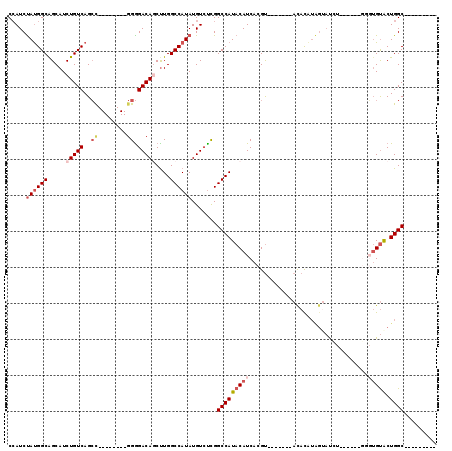
| Location | 20,573,414 – 20,573,509 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.15 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.26 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20573414 95 + 22407834 AGCUUGGCCAUAUGUCUCGGCCAUACAUCACGUACACAUAGUAUCU----UUGUGUGUACUGGCCACACCCCUUGCGAAAGUAAACAGAAAGCGCUAUU .(((((((((((((.......))))......((((((((((.....----))))))))))))))).......((((....)))).....))))...... ( -30.00) >DroSec_CAF1 88171 93 + 1 AGCUUGGCCAUAUGUCUCGGCCAUACAUCACGUACACAUAGUAUCU------GUGUGUGCUGGCCACACCCCCUGCGAAAGUAAACAGAAAGCGCUAUU .(((((((((((((.......))))......((((((((((...))------)))))))))))))........(((....)))......))))...... ( -29.40) >DroSim_CAF1 88846 93 + 1 AGCUUGGCCAUAUGUCUCGGCCAUACAUCACGUACACAUAGUAUCU------GUGUGUACUGGCCACACCCCCUGCGAAAGUAAACAGAAAGCGCUAUU .(((((((((((((.......))))......((((((((((...))------)))))))))))))........(((....)))......))))...... ( -29.40) >DroEre_CAF1 80121 98 + 1 A-CUUGGCCAUAUGUGCCGGCCAUACAUCACGUACAUAUGGUAUGUAUGUACUGCUGUACUGGCCACACCCGUUGCGAAAGUAAGCGGAAAGCGCUAUU .-..(((((....(((..(((((((((.((.((((((((.....)))))))))).)))).))))).)))(((((((....)).)))))...).)))).. ( -37.50) >DroYak_CAF1 82571 93 + 1 AGCCUGGCCAUAUGUCUUGGCCAUACAUCACGUAUAUG-----UGUAUGUACGGGUGUGCUGGCCACACCC-UUGCGAAAGUAAACAGAAAGCGCUAUU ((((((((((.......))))).(((((((((....))-----)).))))).(((((((.....)))))))-((((....))))......)).)))... ( -33.60) >DroPer_CAF1 139074 72 + 1 -----GGCCAUAUGUCUCGGCCAUAUAUCACGUUCCU-------------U-----GGCCUGUC----CUCGAGGCGAAGGCAAACAAAUAGCGCAAUU -----((((.........)))).........((((((-------------(-----.((((...----....)))).))))..)))............. ( -19.50) >consensus AGCUUGGCCAUAUGUCUCGGCCAUACAUCACGUACACAUAGUAUCU______GUGUGUACUGGCCACACCCCUUGCGAAAGUAAACAGAAAGCGCUAUU ......((....(((...(((((........((((((.................)))))))))))........(((....))).)))....))...... (-14.18 = -14.26 + 0.09)

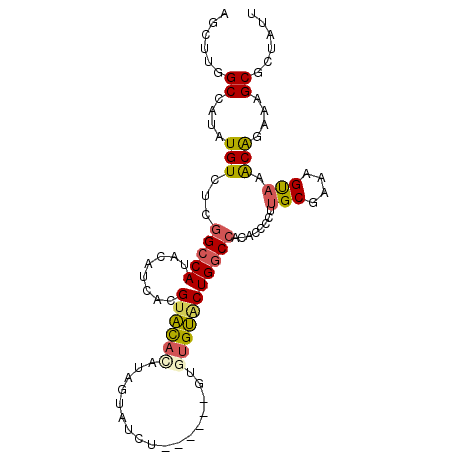
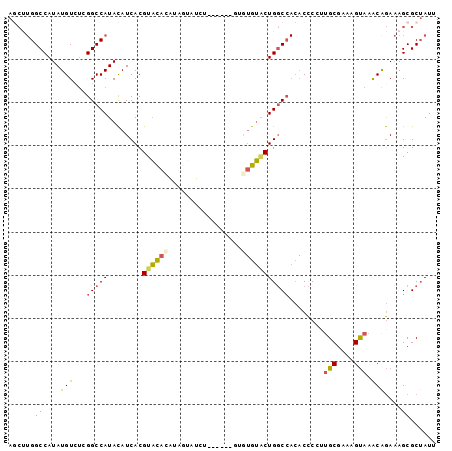
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:44 2006