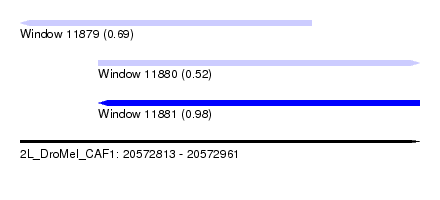
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,572,813 – 20,572,961 |
| Length | 148 |
| Max. P | 0.979852 |

| Location | 20,572,813 – 20,572,921 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 86.44 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -19.06 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20572813 108 - 22407834 CAGACAAACACUGGCCCACUUUGGCCCCACAUCUCC-CCAGAUACUGGACAGCCAACCUGGCCGGCAAGCAGGAGCAACCAUCCCUUAGCCCCAUGUAACCAUGUCCCC ..((((....((((((....(((((.(((.((((..-..))))..)))...)))))...))))))...(((((.((............)).)).))).....))))... ( -28.10) >DroSec_CAF1 87609 100 - 1 CAGACAAACACUGGCCCAGUUUGGCCCCACAUCUCC-CCAGCCACUGGGCAGCCAACCUGGCCGGGAAGCAGGAGCAACCAUCCCUUAGCCCCA--------UGUCCCC ..((((......((((.((.(((((..........(-((((...)))))..))))).))))))(((.....(((.......))).....)))..--------))))... ( -31.00) >DroSim_CAF1 88293 100 - 1 CAGACAAACACUGGCCCAGUUUGGCCCCACAUCUCC-CCAGCCACUGGGCAGCCAACCUGGCCGGGAAGCAGGAGCAACCAUCCCUUAACCCCA--------UGUCCCC ..((((......((((.((.(((((..........(-((((...)))))..))))).))))))(((.....(((.......)))......))).--------))))... ( -30.20) >DroYak_CAF1 81967 99 - 1 CAGUAUAACACUGGCCCAC--UGGCCCCACAUCUACAGCCGCCACUGGGCAAGCAACCUGUCCGGGAAGCAGGAGUAACCAUCCCUUAGCCCCA--------UAUCCCC ((((.....))))(((((.--((((...............)))).)))))..((..(((....)))..)).(((.......)))..........--------....... ( -24.96) >consensus CAGACAAACACUGGCCCACUUUGGCCCCACAUCUCC_CCAGCCACUGGGCAGCCAACCUGGCCGGGAAGCAGGAGCAACCAUCCCUUAGCCCCA________UGUCCCC ............((((......))))....................((((((((.....))).(((.....(((.......))).....)))..........))))).. (-19.06 = -19.25 + 0.19)

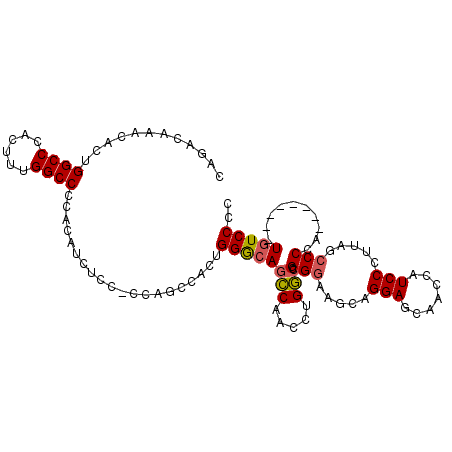
| Location | 20,572,842 – 20,572,961 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -46.32 |
| Consensus MFE | -31.04 |
| Energy contribution | -31.64 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20572842 119 + 22407834 GGUUGCUCCUGCUUGCCGGCCAGGUUGGCUGUCCAGUAUCUGG-GGAGAUGUGGGGCCAAAGUGGGCCAGUGUUUGUCUGGGCAUGACAGCAACUAAGCUGCAGCUGAGCGGGAUUUGAU ......((((((((((((((..(((((.(((((..((..(..(-(.(((..(..((((......)))).)..))).))..)..))))))))))))..))))..)).))))))))...... ( -48.70) >DroSec_CAF1 87630 119 + 1 GGUUGCUCCUGCUUCCCGGCCAGGUUGGCUGCCCAGUGGCUGG-GGAGAUGUGGGGCCAAACUGGGCCAGUGUUUGUCUGGGCAUGACAGCAACUAAGCUGCAGCUGAGCAGGAUUCGAU .((((.(((((((((((((((((((.....)))...)))))))-).(((..(..((((......)))).)..))).....(((.((.((((......)))))))))))))))))..)))) ( -49.30) >DroSim_CAF1 88314 119 + 1 GGUUGCUCCUGCUUCCCGGCCAGGUUGGCUGCCCAGUGGCUGG-GGAGAUGUGGGGCCAAACUGGGCCAGUGUUUGUCUGGGCAUGACAGCAACUAAGCUGCAGCUGAGCAGGAUUUGAU (((((((((..((((((((((((((.....)))...)))))))-))))....)))))..))))....(((..((((.((.(((.((.((((......))))))))).))))))..))).. ( -48.70) >DroEre_CAF1 79650 96 + 1 -GUUACUCCUGCUG-CAGGACAGGUUGCUUGCCCAGUGGC---UGCGGAUG-------------------UGUUUACCUGGGCAUGACAGCAACUAAGCUGCAGCUAAGCGGGAUUUGAU -....(((((((((-(((....((((((((((((((.(..---.(((....-------------------)))...))))))))....)))))))...)))))))..)).)))....... ( -37.00) >DroYak_CAF1 81988 118 + 1 GGUUACUCCUGCUUCCCGGACAGGUUGCUUGCCCAGUGGCGGCUGUAGAUGUGGGGCCA--GUGGGCCAGUGUUAUACUGGCCAUGACAGCAAGUAAACUGCAGCUGAGCGGGAUUUGAU ......((((((((....(.(((.((((((((.((.((((..(..(....)..).))))--.))((((((((...))))))))......)))))))).))))....))))))))...... ( -47.90) >consensus GGUUGCUCCUGCUUCCCGGCCAGGUUGGCUGCCCAGUGGCUGG_GGAGAUGUGGGGCCAAACUGGGCCAGUGUUUGUCUGGGCAUGACAGCAACUAAGCUGCAGCUGAGCGGGAUUUGAU ......((((((((........(((((..(((((((........(.(((((...((((......))))..))))).))))))))...((((......)))))))))))))))))...... (-31.04 = -31.64 + 0.60)



| Location | 20,572,842 – 20,572,961 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -39.63 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.70 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20572842 119 - 22407834 AUCAAAUCCCGCUCAGCUGCAGCUUAGUUGCUGUCAUGCCCAGACAAACACUGGCCCACUUUGGCCCCACAUCUCC-CCAGAUACUGGACAGCCAACCUGGCCGGCAAGCAGGAGCAACC ..........((((.((((((((......)))))..((((.((.......))((((....(((((.(((.((((..-..))))..)))...)))))...)))))))))))..)))).... ( -37.00) >DroSec_CAF1 87630 119 - 1 AUCGAAUCCUGCUCAGCUGCAGCUUAGUUGCUGUCAUGCCCAGACAAACACUGGCCCAGUUUGGCCCCACAUCUCC-CCAGCCACUGGGCAGCCAACCUGGCCGGGAAGCAGGAGCAACC ......((((((((.((((((((......)))).)).))...........((((((.((.(((((..........(-((((...)))))..))))).))))))))).)))))))...... ( -44.00) >DroSim_CAF1 88314 119 - 1 AUCAAAUCCUGCUCAGCUGCAGCUUAGUUGCUGUCAUGCCCAGACAAACACUGGCCCAGUUUGGCCCCACAUCUCC-CCAGCCACUGGGCAGCCAACCUGGCCGGGAAGCAGGAGCAACC ......((((((((.((((((((......)))).)).))...........((((((.((.(((((..........(-((((...)))))..))))).))))))))).)))))))...... ( -44.00) >DroEre_CAF1 79650 96 - 1 AUCAAAUCCCGCUUAGCUGCAGCUUAGUUGCUGUCAUGCCCAGGUAAACA-------------------CAUCCGCA---GCCACUGGGCAAGCAACCUGUCCUG-CAGCAGGAGUAAC- ..........((((.(((((((..((((((((....(((((((.......-------------------........---....)))))))))))).)))..)))-))))..))))...- ( -35.31) >DroYak_CAF1 81988 118 - 1 AUCAAAUCCCGCUCAGCUGCAGUUUACUUGCUGUCAUGGCCAGUAUAACACUGGCCCAC--UGGCCCCACAUCUACAGCCGCCACUGGGCAAGCAACCUGUCCGGGAAGCAGGAGUAACC ......(((((..(((.(((.(((((((.(((.....))).)))).)))....(((((.--((((...............)))).)))))..)))..)))..)))))............. ( -37.86) >consensus AUCAAAUCCCGCUCAGCUGCAGCUUAGUUGCUGUCAUGCCCAGACAAACACUGGCCCACUUUGGCCCCACAUCUCC_CCAGCCACUGGGCAGCCAACCUGGCCGGGAAGCAGGAGCAACC ..........((((.((((((((......)))))..(((((((.........((((......))))......(.......)...)))))))(((.....))).....)))..)))).... (-26.74 = -26.70 + -0.04)

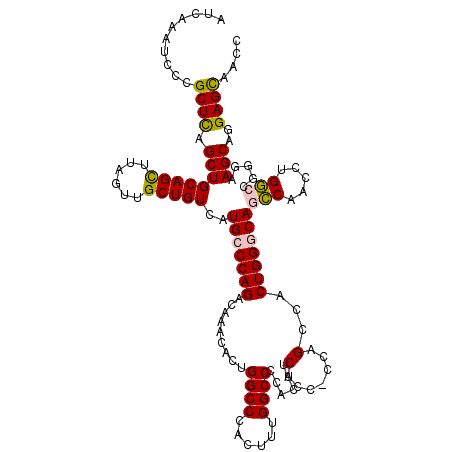
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:41 2006