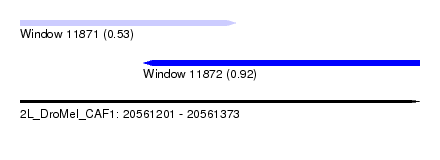
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,561,201 – 20,561,373 |
| Length | 172 |
| Max. P | 0.915053 |

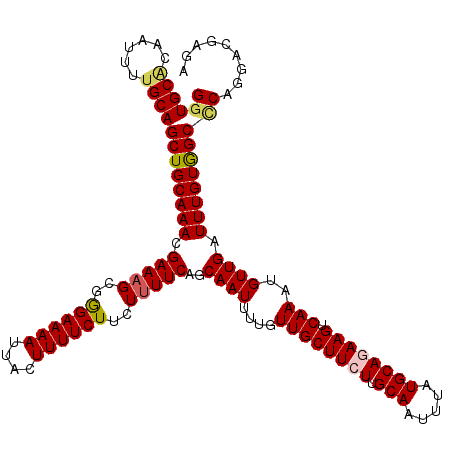
| Location | 20,561,201 – 20,561,294 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.62 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20561201 93 + 22407834 GUGGUCUGUGCCCUGUCCAGGUGUUCCGGAGCCUGG-ACUCCGAGUCCCUGCU----------------CCCUCGUCCUGAGCCACAAAUCAACAUUUGACUUCUGCAUA .((((.((((....(..((((((....(((((..((-((.....))))..)))----------------))..)).))))..))))).)))).................. ( -27.70) >DroSec_CAF1 83507 93 + 1 GUGGUCUCUGCCCUGUCCAGGUGUUCCGGAGCCUGG-ACUCCGAGUCCCUGCU----------------CUCUCGUCCUGGGCUACAAAUCAACAUUUGACUUCUGCAUA (..(..........(((((((((....(((((..((-((.....))))..)))----------------))..)).)))))))..(((((....)))))....)..)... ( -29.00) >DroSim_CAF1 84232 93 + 1 GUGGUCUCUGCCCUGUCCAGGUGUUCCGGAGCCUGG-ACUCCGACUCCCUGCU----------------CCCUCGUCCUGGGCCACAAAUCAACAUUUGACUUCUGCAUA (..(..........(((((((((....(((((..((-(.......)))..)))----------------))..)).)))))))..(((((....)))))....)..)... ( -26.50) >DroEre_CAF1 75623 108 + 1 GUGGUCUCUGCCCUGUCCAGGUGUUCCGGAGCCUGG-ACUCCGAGUCCUGGCUCCUCAGCUCCACAGCUCC-CCGUCCUGGGCCACAAAUCAACAUUUGACUUCUGCAUA (..(..........(((((((((....((((((.((-((.....)))).))))))..((((....))))..-.)).)))))))..(((((....)))))....)..)... ( -36.90) >DroYak_CAF1 77761 108 + 1 GUGGUCUCUGC-CUGUCCAGGUGUUCCGCAGCCUGG-ACUCCAAGUCCUGGCUCCGCAGCUCCGCAGCUCCCCCGUCCUGGGCCACAAAUCAACAUUUGACUUCUGCAUA .(((.(.....-..).)))((.(((.((.((((.((-((.....)))).)))).)).))).))((((....(((.....)))...(((((....)))))....))))... ( -33.30) >DroAna_CAF1 76162 78 + 1 GUGGUCCCUG----GUCCAGGUGCUGCAGAUUCUGGCACUC----------------------------CUCUGGUCCUGGGCCACAAAUCAACAUUUGACUUUUGCAUA (((((((..(----(.(((((((((.........)))))..----------------------------..)))).)).)))))))........................ ( -27.30) >consensus GUGGUCUCUGCCCUGUCCAGGUGUUCCGGAGCCUGG_ACUCCGAGUCCCUGCU________________CCCUCGUCCUGGGCCACAAAUCAACAUUUGACUUCUGCAUA (((((((.......((((((((........)))))).))....(((....)))..........................)))))))........................ (-17.41 = -17.62 + 0.21)

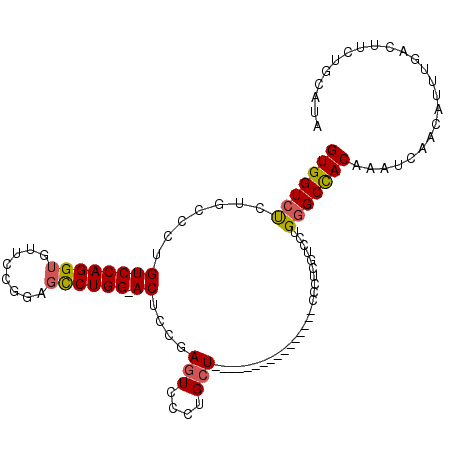
| Location | 20,561,254 – 20,561,373 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -28.78 |
| Energy contribution | -29.03 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20561254 119 - 22407834 GGUGCACAAU-UUGCAGCUGCAAACGAAAGCGGGAAAAUUACUUUUCUUCUUUUCAGCAAUUUUGUUGCUUCUUGCAAUUUAUGCAGAAGUCAAAUGUUGAUUUGUGGCUCAGGACGAGG .........(-(((.(((..((((..((((.((((((.....)))))).))))((((((..((((..((((((.(((.....)))))))))))))))))))))))..)))))))...... ( -31.60) >DroPse_CAF1 129585 113 - 1 GGUGC-------UGCAGCUGCAAACGAAAGCGAGAAAAUUACUUUUCUUCUUUUCAGCAAUUUUGUUGCUUCUUGCAAUUUAUGCAAAAGUCAAAUGUUGAUUUGUGGCCCAGGACCACA (((.(-------((..((..((((..((((.(((((((....)))))))))))((((((..((((..((((.(((((.....)))))))))))))))))))))))..)).))).)))... ( -35.00) >DroSec_CAF1 83560 120 - 1 GGUGCACAAUUUUGCAGCUGCAAACGAAAGCGGGAAAAUUACUUUUCUUCUUUUCAGCAAUUUUGUUGCUUCUUGCAAUUUAUGCAGAAGUCAAAUGUUGAUUUGUAGCCCAGGACGAGA ((((((......))))((((((((..((((.((((((.....)))))).))))((((((..((((..((((((.(((.....)))))))))))))))))))))))))))))......... ( -33.50) >DroEre_CAF1 75692 119 - 1 GGUGCACAAUUUUGCAGCUGCAAACGAAAGCGGGAAAAUUACUUUUCUUCUUUUCAGCAAUUUUGUUGCUUCUUGCAAUUUAUGCAGAAGUCAAAUGUUGAUUUGUGGCCCAGGACGG-G ((((((......))))((..((((..((((.((((((.....)))))).))))((((((..((((..((((((.(((.....)))))))))))))))))))))))..)))).......-. ( -32.30) >DroYak_CAF1 77829 120 - 1 GGUGCGCAGUUUUGCAGCUGCAAACGAAAGCGGGAAAAUUACUUUUCUUCUCUUCAGCAAUUUUGUUGCUUCUUGCAAUUUAUGCAGAAGUCAAAUGUUGAUUUGUGGCCCAGGACGGGG ((.(((((((......)))))..(((((((.((((((.....)))))).))..((((((..((((..((((((.(((.....)))))))))))))))))))))))).))))......... ( -32.40) >DroAna_CAF1 76200 118 - 1 GUUGCAC--UUCUGCAGCUGCAAACGAAAGCGAGAAAAUUACUUUUCUUCUUUUCAGCAAUUUUGUUGCUUCUUGCAAUUUAUGCAAAAGUCAAAUGUUGAUUUGUGGCCCAGGACCAGA .......--(((((..((..((((..((((.(((((((....)))))))))))((((((..((((..((((.(((((.....)))))))))))))))))))))))..)).)))))..... ( -29.70) >consensus GGUGCACAAUUUUGCAGCUGCAAACGAAAGCGGGAAAAUUACUUUUCUUCUUUUCAGCAAUUUUGUUGCUUCUUGCAAUUUAUGCAGAAGUCAAAUGUUGAUUUGUGGCCCAGGACGAGA ((((((......))))((((((((.(((((..((((((....))))))..)))))..((((....((((((((.(((.....)))))))).)))..)))).))))))))))......... (-28.78 = -29.03 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:33 2006