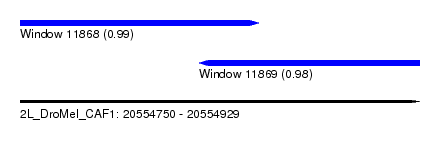
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,554,750 – 20,554,929 |
| Length | 179 |
| Max. P | 0.985422 |

| Location | 20,554,750 – 20,554,857 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 73.70 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -15.24 |
| Energy contribution | -14.47 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
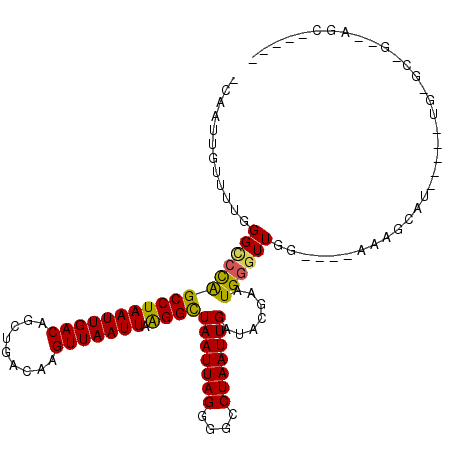
>2L_DroMel_CAF1 20554750 107 + 22407834 GCAAUUGUUUUGGGCCUGGCCUAAUUGACAGCUGGCAAGUUAAUUAAGGCUAAUUAGUGGCCUAAUUGAUACGAAUGGGUUGU----AAAGUAUGGCUGUGAGCUGGGAGC----- ((.......((((((...))))))....(((((.((..((((((((.(((((.....))))))))))))).......(((..(----(...))..))))).)))))...))----- ( -31.50) >DroPse_CAF1 120999 102 + 1 -GAAUUGUUUUUGGUCCAGCCUAAUUGACAGCUGACAAGUUAAUUAAGGUUAAUUAGGAUGCUAAUUGAUACGAAUGGGUUGG-------GCU------UGGGCCGAUAGCCAGUU -.(((((((.((((((((((((((((...((((....)))).(((...(((((((((....)))))))))...))).))))))-------)).------)))))))).)).))))) ( -33.60) >DroEre_CAF1 68609 97 + 1 GCAAUUGUUU-GGGCCCGGCCUAAUUGACAGCUGGCAAGUUAAUUAGGGCUAAUUAGUGGCCUAAUUGAUACGAAUGGCUUGC----AAGGCAUGGGUG---------AGC----- ..........-..(((((((((...((..(((((....((((((((((.(........).)))))))))).....)))))..)----))))).))))).---------...----- ( -30.30) >DroYak_CAF1 71735 107 + 1 GCAAUUGUUUAGGGCCUGGCCUAAUUGACAGCUGGCAAGUUAAUUAAGGCUAAUUAGUGGCCUAAUUGAUACGAAUGGGUUGC----AAAGUAUGGGUGUGAGAUAGGAGC----- ((..((((((.(.((((((((((((((((.........))))))).))))).....(..(((((.(((...))).)))))..)----.......)))).).))))))..))----- ( -31.20) >DroAna_CAF1 69970 93 + 1 -GAAUUGAUUUGGGUGCAGCCUAAUUGACAGCUGACAAGUUAAUUAGGGUUAAUUAGGGCUCUAAUUGAUACGAAUGGGUUGGCCUUAAAAUGC---------------------- -.......((((((..(((((((.(((.(....)....(((((((((((((......))))))))))))).))).)))))))..))))))....---------------------- ( -25.20) >DroPer_CAF1 126518 102 + 1 -GAAUUGUUUUUGGUCCAGCCUAAUUGACAGCUGACAAGUUAAUUAAGGUUAAUUAGGAUGCUAAUUGAUACGAAUGGGUUGG-------GCU------UGGGCCGAUAGCCAGUU -.(((((((.((((((((((((((((...((((....)))).(((...(((((((((....)))))))))...))).))))))-------)).------)))))))).)).))))) ( -33.60) >consensus _CAAUUGUUUUGGGCCCAGCCUAAUUGACAGCUGACAAGUUAAUUAAGGCUAAUUAGGGGCCUAAUUGAUACGAAUGGGUUGG____AAAGCAU_____UG_GC_G__AGC_____ ............(((((((((((((((((.........))))))).))))(((((((....))))))).......))))))................................... (-15.24 = -14.47 + -0.78)

| Location | 20,554,830 – 20,554,929 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -16.06 |
| Energy contribution | -15.12 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
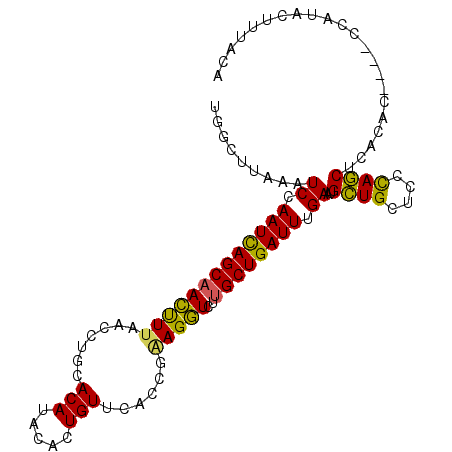
>2L_DroMel_CAF1 20554830 99 - 22407834 UGGCUUAAAUCCAAUCAGCAACUUAAACCUUCACAUACACUGUUUACCGAAGGUCUUGCUGAUUUGAAUGCUGCUCCCAGCUCACAG----CCAUACUUUACA (((((....((.(((((((((.....(((((((((.....))).....)))))).))))))))).))..((((....))))....))----)))......... ( -27.10) >DroSec_CAF1 77581 103 - 1 AGGCUUAAAUCCAAUCAGCAACUUUAACCAGCACAUACAAUGUUCACCGAAGGUCUUGCUGAUUUGAAUGCUGCUCCCAGCUCACACCCACCCAUAUUUUACA .((......((.(((((((((((((....((((.......)))).....)))))..)))))))).))..((((....))))......)).............. ( -19.40) >DroSim_CAF1 78266 103 - 1 UGGCUUAAAUCCAAUCAGCAACUUUAACCUUCACAUAAACUGUUCACCGAAGGUCUUGCUGAUUUGAAUGCUGCUCCCAGCUCACACCCACCCAUAUUUUACA (((......((.(((((((((.....(((((((((.....))).....)))))).))))))))).))..((((....))))..........)))......... ( -22.40) >DroYak_CAF1 71815 99 - 1 UUGGAUAAGUCCAAUUAGCAAGCUUAACCUGCACAUACACUGUUUAUGAGAGCUCUGGCUGAUUUGAAUGGUGCUCCUAUCUCACAC----CCAUACUUUGCA (((((....)))))...((((((.......)).....(((((((((....(((....)))....)))))))))..............----.......)))). ( -19.70) >consensus UGGCUUAAAUCCAAUCAGCAACUUUAACCUGCACAUACACUGUUCACCGAAGGUCUUGCUGAUUUGAAUGCUGCUCCCAGCUCACAC____CCAUACUUUACA .........((.(((((((((((((.......(((.....)))......)))))..)))))))).))..((((....))))...................... (-16.06 = -15.12 + -0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:30 2006