| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,548,744 – 20,548,840 |
| Length | 96 |
| Max. P | 0.997280 |

| Location | 20,548,744 – 20,548,840 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -37.01 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.85 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.816098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
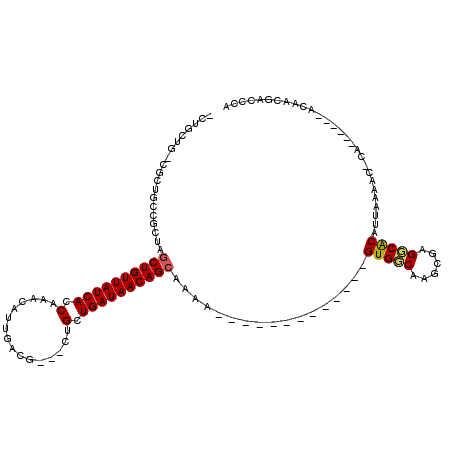
>2L_DroMel_CAF1 20548744 96 + 22407834 UGGGUCGUUGU------UG-GUUUUAAUGCGACUCGCUUGCCAC-------------UUUUGCUGUUAUCAGCAG---CGUCAAUGUUUGGUGAUAACAGCCAGCGGCAGCG-CAGCAGC .(((((((...------..-........)))))))(((((((.(-------------(...((((((((((.(((---((....)).))).)))))))))).)).))))).)-)...... ( -37.12) >DroPse_CAF1 113637 117 + 1 UGGGUCGUUGUUGUUGUUG-GUUUUAAUGUGCCUCCAUUGGCACACUCUGCCACACGCUUUGCUGUUAUCAGCAGCGGCGUCAAUGUUUGGUGAUAACAGCUCAAGGCAGCG-CAGCAG- ......(((((.(((((((-((.....((((((......))))))....)))).))(((((((((((((((.(((((.......)).))).))))))))))..)))))))))-))))..- ( -43.40) >DroEre_CAF1 62857 95 + 1 UGGGUCGUUGU------UG-CUUUUAAUGCGACUCGCUUGCCAC-------------UUUUGCUGUUAUCAGCUG---CGCCAAUGUUUGCUGAUAACAGCCAGCGGCAUCG-CAGCUG- .(((((((...------..-........)))))))((.((((.(-------------(...((((((((((((.(---((....)))..)))))))))))).)).))))..)-).....- ( -39.12) >DroYak_CAF1 65733 86 + 1 UGGGUCGUUGU------UG-GUUUUAAUGCGACUCGCUUGCCAC-------------UUUUGCUGUUAUCAGCUG---CGCCAAUGUUUGGUGAUAACAGCCAGCGGCA----------- .(((((((...------..-........)))))))...((((.(-------------(...((((((((((.(.(---((....)))..).)))))))))).)).))))----------- ( -31.42) >DroAna_CAF1 63673 98 + 1 UGGGUCGUUGU------UGGGUUUUAAUGUGCCUCGGUUGCCAC-------------UUUUUCUGUUAUCAGCAG---CGUCAAUGUUUGGUGAUAACAGCUACAGGCAGAGGCUGUGGC .(((((((((.------.......))))).)))).....(((((-------------..((((((((((((.(((---((....)).))).))))))).((.....)))))))..))))) ( -28.90) >DroPer_CAF1 119170 114 + 1 UGGGUCGUUGU---UGUUG-GUUUUAAUGUGCCUCCAUUGGCACACUCUGCCACACGCUUUGCUGUUAUCAGCAGCGGCGUCAAUGUUUGGUGAUAACAGCUCAAGGCAGCG-CAGCAG- ......(((((---.((((-((.....((((((......))))))....)))).))(((((((((((((((.(((((.......)).))).))))))))))..)))))...)-))))..- ( -42.10) >consensus UGGGUCGUUGU______UG_GUUUUAAUGCGACUCGCUUGCCAC_____________UUUUGCUGUUAUCAGCAG___CGUCAAUGUUUGGUGAUAACAGCCAGAGGCAGCG_CAGCAG_ .(((.(((((..............)))))...)))..........................((((((((((.(................).))))))))))................... (-14.68 = -14.85 + 0.17)


| Location | 20,548,744 – 20,548,840 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -35.14 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.16 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20548744 96 - 22407834 GCUGCUG-CGCUGCCGCUGGCUGUUAUCACCAAACAUUGACG---CUGCUGAUAACAGCAAAA-------------GUGGCAAGCGAGUCGCAUUAAAAC-CA------ACAACGACCCA ((.(((.-((((((((((.((((((((((.((..........---.)).))))))))))...)-------------))))).))))))).))........-..------........... ( -37.40) >DroPse_CAF1 113637 117 - 1 -CUGCUG-CGCUGCCUUGAGCUGUUAUCACCAAACAUUGACGCCGCUGCUGAUAACAGCAAAGCGUGUGGCAGAGUGUGCCAAUGGAGGCACAUUAAAAC-CAACAACAACAACGACCCA -((((..-(((.((.((..((((((((((.((..............)).)))))))))).)))))))..))))((((((((......)))))))).....-................... ( -40.44) >DroEre_CAF1 62857 95 - 1 -CAGCUG-CGAUGCCGCUGGCUGUUAUCAGCAAACAUUGGCG---CAGCUGAUAACAGCAAAA-------------GUGGCAAGCGAGUCGCAUUAAAAG-CA------ACAACGACCCA -..(.((-(..(((((((.((((((((((((...........---..))))))))))))...)-------------)))))).(((...))).......)-))------.)......... ( -37.62) >DroYak_CAF1 65733 86 - 1 -----------UGCCGCUGGCUGUUAUCACCAAACAUUGGCG---CAGCUGAUAACAGCAAAA-------------GUGGCAAGCGAGUCGCAUUAAAAC-CA------ACAACGACCCA -----------(((((((.(((((((((((((.....)))..---....))))))))))...)-------------))))))...(.((((.........-..------....)))).). ( -29.56) >DroAna_CAF1 63673 98 - 1 GCCACAGCCUCUGCCUGUAGCUGUUAUCACCAAACAUUGACG---CUGCUGAUAACAGAAAAA-------------GUGGCAACCGAGGCACAUUAAAACCCA------ACAACGACCCA ......(((((((((.....(((((((((.((..........---.)).))))))))).....-------------..))))...))))).............------........... ( -25.40) >DroPer_CAF1 119170 114 - 1 -CUGCUG-CGCUGCCUUGAGCUGUUAUCACCAAACAUUGACGCCGCUGCUGAUAACAGCAAAGCGUGUGGCAGAGUGUGCCAAUGGAGGCACAUUAAAAC-CAACA---ACAACGACCCA -((((..-(((.((.((..((((((((((.((..............)).)))))))))).)))))))..))))((((((((......)))))))).....-.....---........... ( -40.44) >consensus _CUGCUG_CGCUGCCGCUAGCUGUUAUCACCAAACAUUGACG___CUGCUGAUAACAGCAAAA_____________GUGGCAAGCGAGGCACAUUAAAAC_CA______ACAACGACCCA ...................((((((((((.(................).)))))))))).................(((((......)))))............................ (-18.41 = -18.16 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:27 2006