| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,534,249 – 20,534,347 |
| Length | 98 |
| Max. P | 0.702387 |

| Location | 20,534,249 – 20,534,347 |
|---|---|
| Length | 98 |
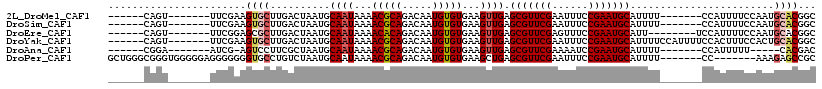
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.10 |
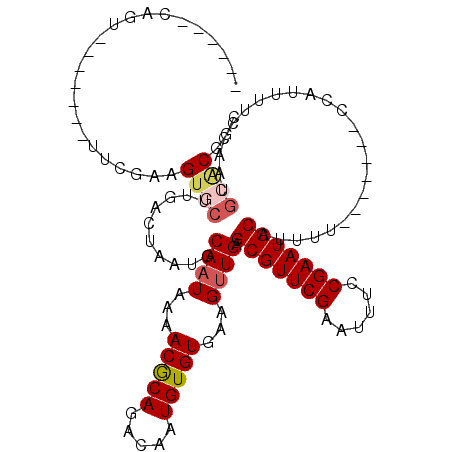
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -19.30 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20534249 98 - 22407834 ------CAGU-------UUCGAAGUGCUUGACUAAUGCAAUAAAACGCAGACAAUGUGUGAAGUUGAGCGUUCGAAUUUCCGAAUGCAUUUU-------CCAUUUUCCAAUGCACGGC ------....-------......(((((((...((((((((...(((((.....)))))...)))).(((((((......))))))).....-------.))))...))).))))... ( -26.10) >DroSim_CAF1 56919 98 - 1 ------CAGU-------UUCGAAGUGCUUGACUAAUGCAAUAAAACGCAGACAAUGUGUGAAGUUGAGCGUUCGAAUUUCCGAAUGCAUUUU-------CCAUUUUCCAAUGCACGGC ------....-------......(((((((...((((((((...(((((.....)))))...)))).(((((((......))))))).....-------.))))...))).))))... ( -26.10) >DroEre_CAF1 48559 97 - 1 ------CAGU-------UUCGGAGCGCUUGACUAAUGCAAUAAAACACAGACAAUGUGUGAAGUUGAGCGUUCGAGUUUCCGAAUGCAUU--------UCCAUUUUCCAAUGCACGGC ------....-------((((.((((((..(((...........(((((.....)))))..)))..))))))))))...(((..((((((--------..........))))))))). ( -26.62) >DroYak_CAF1 50966 105 - 1 ------CAGU-------UUCGAAGUGCUUGACUAAUGCAAUAAAACGCAGACAAUGUGUGAAGUUGAGCGUUCGAAUUUCCGAAUGCAUUUUCCAUUUUCCACUUUCCACUGCACGGC ------((((-------...((((((...((..((((((((...(((((.....)))))...)))).(((((((......)))))))......)))).))))))))..))))...... ( -25.30) >DroAna_CAF1 49666 92 - 1 ------CGGA-------AUCG-AGUCCUUCGCUAAUGCAAUAAAACGCAGACAAUGUGUGAAGUUGAGCGUUCGAAAAUCCGAAUGCAUUUU-------CCAUUUUU-----CACGAC ------.(((-------((((-(...((((((....))......(((((.....)))))))))))))(((((((......)))))))...))-------))......-----...... ( -24.70) >DroPer_CAF1 99175 104 - 1 GCUGGGCGGGUGGGGGAGGGGGGGUGCCUGUCUAAUGCAAUAAAACGCAGACAAUGUGUGAAGCUGAGCGUUCGAAUUUCCGAAUGCAUUUU-------CC-------AAAGAGCCGC ....(((...(((..(((((.((....)).)))...((......(((((.....)))))...))...(((((((......))))))).))..-------))-------)....))).. ( -31.60) >consensus ______CAGU_______UUCGAAGUGCUUGACUAAUGCAAUAAAACGCAGACAAUGUGUGAAGUUGAGCGUUCGAAUUUCCGAAUGCAUUUU_______CCAUUUUCCAAUGCACGGC .......................((((..........((((...(((((.....)))))...)))).(((((((......)))))))........................))))... (-19.30 = -19.88 + 0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:17 2006