
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,531,573 – 20,531,826 |
| Length | 253 |
| Max. P | 0.944767 |

| Location | 20,531,573 – 20,531,668 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
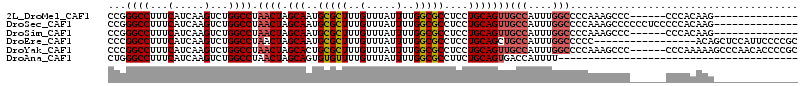
>2L_DroMel_CAF1 20531573 95 - 22407834 CCGGGCCUUUCAUCAAGUCUGGCCUAACUAGCAAUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGUUGCCAUUUGGCCCCAAAGCCC------CCCACAAG-------------- ..((((.............((((..((((.(((..(((((..(.....)..)))))....)))))))))))((((....))))))))------........-------------- ( -26.00) >DroSec_CAF1 52809 101 - 1 CCGGGCCUUUCAUCAAGUCUGGCCUAACUAGCAAUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGUUGCCAUUUGGCCCCAAAGCCCCCCUCCCCCACAAG-------------- ..((((.............((((..((((.(((..(((((..(.....)..)))))....)))))))))))((((....))))))))..............-------------- ( -26.00) >DroSim_CAF1 54136 95 - 1 CCGGGCCUUUCAUCAAGUCUGGCCUAACUAGCAAUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGUUGCCAUUUGGCCCCAAAGCCC------CCCACAAG-------------- ..((((.............((((..((((.(((..(((((..(.....)..)))))....)))))))))))((((....))))))))------........-------------- ( -26.00) >DroEre_CAF1 45943 98 - 1 CCCGGCCUUUCAUCAAGUCUGGCCUAACUAGCAAUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGCUGCCAUUUGGCCCCC-----------------ACAGCUCCAUUCCCCGC ...((((...(.....)...))))......(((..(((((..(.....)..)))))....)))(((((((....)))....-----------------..))))........... ( -21.70) >DroYak_CAF1 48148 109 - 1 CCCGGCCUUUCAUCAAGUCUGGCCUAACUAGCACUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGUUGCCAUUUGGCCCCAAAGCCC------CCCAAAAAGCCCAACACCCCGC ...(((.............((((..((((.(((..(((((..(.....)..)))))....)))))))))))...(((......))).------........)))........... ( -24.00) >DroAna_CAF1 45440 75 - 1 CUGGGCCUUUCAUCAAGUCUGGCCUAACUAGCAGUGUGUUUUGUUUAUUUUGGCGCCUUCUGCAGUGACCAUUUU---------------------------------------- .((((((...(.....)...))))))(((.((((.(((((..(.....)..)))))...))))))).........---------------------------------------- ( -18.80) >consensus CCGGGCCUUUCAUCAAGUCUGGCCUAACUAGCAAUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGUUGCCAUUUGGCCCCAAAGCCC______CCCACAAG______________ ...((((...(.....)...)))).((((.(((..(((((..(.....)..)))))....)))))))(((....)))...................................... (-17.96 = -18.52 + 0.56)

| Location | 20,531,593 – 20,531,687 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.16 |
| Mean single sequence MFE | -31.79 |
| Consensus MFE | -27.03 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20531593 94 + 22407834 CAAAUGGCAACUGCAGGAGGCGCCAAAAUAAACAAAGCGCAUUGCUAGUUAGGCCAGACUUGAUGAAAGGCCCGGGAAAGCUCCUGCCGGCCGG----------------- (((.((((.(((((((...((((.............)))).)))).)))...))))...)))......(((((((((....)))))..))))..----------------- ( -31.72) >DroSec_CAF1 52835 94 + 1 CAAAUGGCAACUGCAGGAGGCGCCAAAAUAAACAAAGCGCAUUGCUAGUUAGGCCAGACUUGAUGAAAGGCCCGGGAAAACUCCUGCCGGCCGG----------------- (((.((((.(((((((...((((.............)))).)))).)))...))))...)))......(((((((((....)))))..))))..----------------- ( -31.72) >DroSim_CAF1 54156 94 + 1 CAAAUGGCAACUGCAGGAGGCGCCAAAAUAAACAAAGCGCAUUGCUAGUUAGGCCAGACUUGAUGAAAGGCCCGGGAAAACUCCUGCCGGCCGG----------------- (((.((((.(((((((...((((.............)))).)))).)))...))))...)))......(((((((((....)))))..))))..----------------- ( -31.72) >DroEre_CAF1 45966 94 + 1 CAAAUGGCAGCUGCAGGAGGCGCCAAAAUAAACAAAGCGCAUUGCUAGUUAGGCCAGACUUGAUGAAAGGCCGGGGAAAACUCCUGCCGGCCGG----------------- (((.((((.(((((((...((((.............)))).)))).)))...))))...)))......(((((((((....)))..))))))..----------------- ( -34.22) >DroYak_CAF1 48182 94 + 1 CAAAUGGCAACUGCAGGAGGCGCCAAAAUAAACAAAGCGCAGUGCUAGUUAGGCCAGACUUGAUGAAAGGCCGGGGAAAACUCCUGCCGGCCGG----------------- (((.((((.((((((....((((.............))))..))).)))...))))...)))......(((((((((....)))..))))))..----------------- ( -31.92) >DroAna_CAF1 45440 111 + 1 AAAAUGGUCACUGCAGAAGGCGCCAAAAUAAACAAAACACACUGCUAGUUAGGCCAGACUUGAUGAAAGGCCCAGGAAAAGUCCUGCCGGGCAGAGAGAUCUGCGGAGCAG ....((((..((.....))..))))................(((((.....((((.............))))(((((....)))))((..(((((....)))))))))))) ( -29.42) >consensus CAAAUGGCAACUGCAGGAGGCGCCAAAAUAAACAAAGCGCAUUGCUAGUUAGGCCAGACUUGAUGAAAGGCCCGGGAAAACUCCUGCCGGCCGG_________________ ....((((.(((((((...((((.............)))).)))).)))...))))............(((((((((....)))))..))))................... (-27.03 = -27.50 + 0.47)

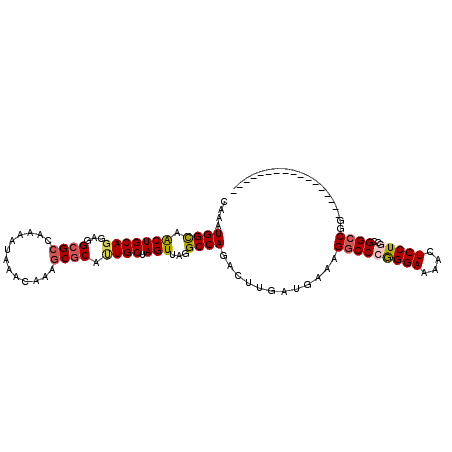

| Location | 20,531,593 – 20,531,687 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.16 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -28.36 |
| Energy contribution | -28.45 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20531593 94 - 22407834 -----------------CCGGCCGGCAGGAGCUUUCCCGGGCCUUUCAUCAAGUCUGGCCUAACUAGCAAUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGUUGCCAUUUG -----------------..(((..(((((((((.....(((((...(.....)...)))))....)))...(((((..(.....)..))))).))))))....)))..... ( -33.60) >DroSec_CAF1 52835 94 - 1 -----------------CCGGCCGGCAGGAGUUUUCCCGGGCCUUUCAUCAAGUCUGGCCUAACUAGCAAUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGUUGCCAUUUG -----------------..(((..(((((((.......(((((...(.....)...)))))..........(((((..(.....)..))))))))))))....)))..... ( -32.90) >DroSim_CAF1 54156 94 - 1 -----------------CCGGCCGGCAGGAGUUUUCCCGGGCCUUUCAUCAAGUCUGGCCUAACUAGCAAUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGUUGCCAUUUG -----------------..(((..(((((((.......(((((...(.....)...)))))..........(((((..(.....)..))))))))))))....)))..... ( -32.90) >DroEre_CAF1 45966 94 - 1 -----------------CCGGCCGGCAGGAGUUUUCCCCGGCCUUUCAUCAAGUCUGGCCUAACUAGCAAUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGCUGCCAUUUG -----------------..(((..(((((((........((((...(.....)...))))...........(((((..(.....)..))))))))))))....)))..... ( -31.80) >DroYak_CAF1 48182 94 - 1 -----------------CCGGCCGGCAGGAGUUUUCCCCGGCCUUUCAUCAAGUCUGGCCUAACUAGCACUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGUUGCCAUUUG -----------------..(((..(((((((........((((...(.....)...))))...........(((((..(.....)..))))))))))))....)))..... ( -31.80) >DroAna_CAF1 45440 111 - 1 CUGCUCCGCAGAUCUCUCUGCCCGGCAGGACUUUUCCUGGGCCUUUCAUCAAGUCUGGCCUAACUAGCAGUGUGUUUUGUUUAUUUUGGCGCCUUCUGCAGUGACCAUUUU (((((..(((((....)))))..))))).........((((((...(.....)...))))))(((.((((.(((((..(.....)..)))))...)))))))......... ( -32.30) >consensus _________________CCGGCCGGCAGGAGUUUUCCCGGGCCUUUCAUCAAGUCUGGCCUAACUAGCAAUGCGCUUUGUUUAUUUUGGCGCCUCCUGCAGUUGCCAUUUG ...................(((..(((((((........((((...(.....)...))))...........(((((..(.....)..))))))))))))....)))..... (-28.36 = -28.45 + 0.09)

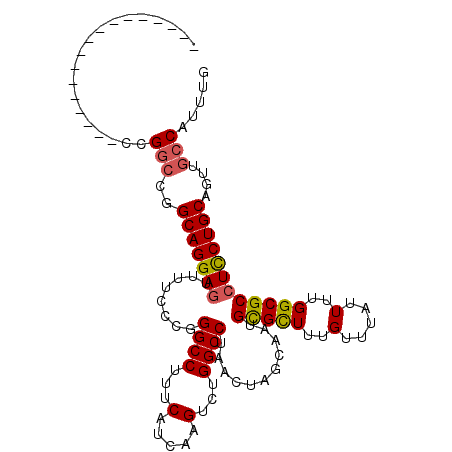

| Location | 20,531,687 – 20,531,795 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -24.77 |
| Energy contribution | -25.13 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20531687 108 - 22407834 UAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGGCCGCCGCC---AAAUUUUCCC------- ...((((((.(((((....(((((((((((..(((....((.......))....)))..)))))).)))))((((.((.....))..)))).))))))---))))).....------- ( -31.60) >DroSec_CAF1 52929 108 - 1 UAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGGCCGCCGCC---AAAUUUUCCC------- ...((((((.(((((....(((((((((((..(((....((.......))....)))..)))))).)))))((((.((.....))..)))).))))))---))))).....------- ( -31.60) >DroSim_CAF1 54250 108 - 1 UAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGGCCGCCGCC---AAAUUUUCCC------- ...((((((.(((((....(((((((((((..(((....((.......))....)))..)))))).)))))((((.((.....))..)))).))))))---))))).....------- ( -31.60) >DroEre_CAF1 46060 108 - 1 UAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUCUCGGCCGCCGCC---AAAUUUUCCC------- ...((((((.(((((..(((((((((((((..(((....((.......))....)))..))))))......(.....)......))))))).))))))---))))).....------- ( -33.10) >DroYak_CAF1 48276 108 - 1 UAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGUCCGCCGCC---AAAUUUUCCC------- ...((((((.(((((....(((((((((((..(((....((.......))....)))..)))))).)))))(..((.......))..)....))))))---))))).....------- ( -31.20) >DroAna_CAF1 45551 117 - 1 UAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAUCCAAC-UCGGUCUCCUCCCGAAAAUUGUCCUCUUUCCU ..........(..((.((..((((((((((..(((....((.......))....)))..))))))))))))))..)((((.....-((((.......)))).....))))........ ( -31.00) >consensus UAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAUCCAGUUUCGGCCGCCGCC___AAAUUUUCCC_______ ......(((..(((((....((((((((((..(((....((.......))....)))..))))))))))....))).))..)))....(((....))).................... (-24.77 = -25.13 + 0.36)

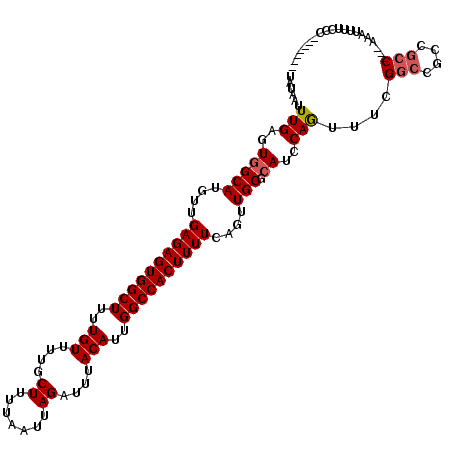
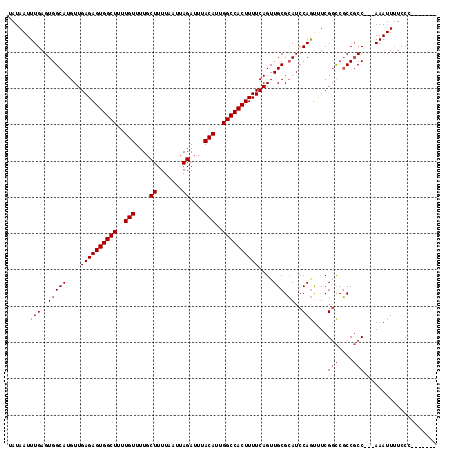
| Location | 20,531,715 – 20,531,826 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20531715 111 - 22407834 UCAACUG-GG-------CGGAG-GCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAU .((((((-((-------((...-.....))))((((((.(((.....)))...)))))).((((((((((..(((....((.......))....)))..))))))))))))))))..... ( -34.50) >DroPse_CAF1 92320 110 - 1 NN--NNN-NN------NNNUGG-GCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAU ..--...-..------......-.....(((.((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...)))... ( -27.20) >DroSec_CAF1 52957 112 - 1 UCCACAG-GG-------CGGAGGGCCCGCGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAU ......(-((-------(.....))))((((.((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...)))).. ( -38.70) >DroEre_CAF1 46088 107 - 1 ----CCG-GA-------CGGAG-GCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAU ----((.-..-------.))..-((.((.(((((((((.(((.....)))...)))))).((((((((((..(((....((.......))....)))..)))))))))).))))).)).. ( -27.70) >DroAna_CAF1 45588 119 - 1 UUGGCCAGGACUCUCCUUGCAG-GCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAU ..(((((((.....)))....)-)))..(((.((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...)))... ( -35.80) >DroPer_CAF1 97269 110 - 1 UC--CAG-UG------CAGUGG-GCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGGAU ((--(((-((------((...(-((....)))...)))))).........(..((.((..((((((((((..(((....((.......))....)))..))))))))))))))..)))). ( -36.00) >consensus UC__CAG_GG_______CGGAG_GCCCACGCUAUAUGCAUUAUAAUUUGAGUGGCAUGUUGAGAGUGGCUUUUGUUUUGCUUUUAAUUAGAUUUACAUUGGCCACUUUUCAGUUGCGCAU ............................(((.((((((.(((.....)))...))))))(((((((((((..(((....((.......))....)))..)))))).)))))...)))... (-26.70 = -26.70 + -0.00)

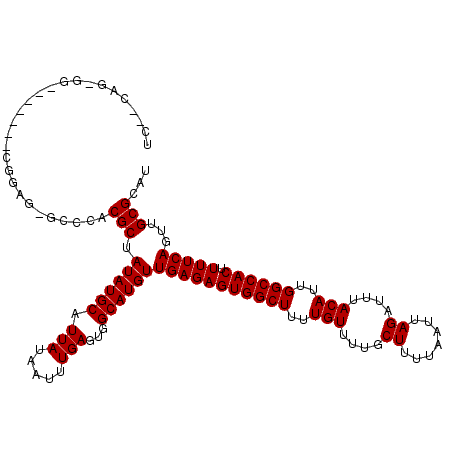
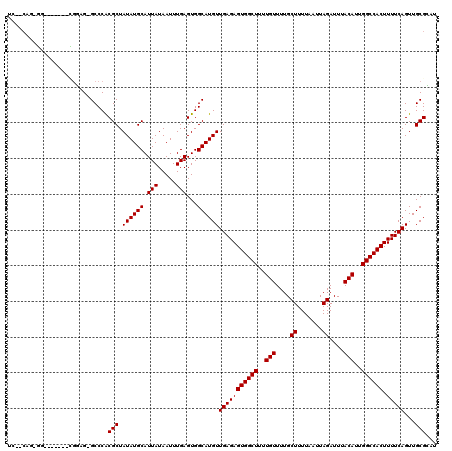
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:13 2006