
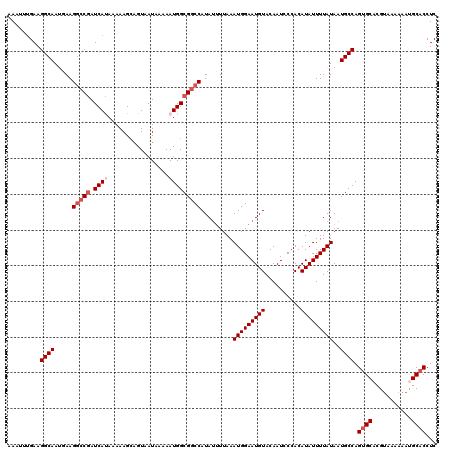
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,528,424 – 20,528,623 |
| Length | 199 |
| Max. P | 0.978969 |

| Location | 20,528,424 – 20,528,544 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -18.39 |
| Energy contribution | -19.59 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20528424 120 + 22407834 AAAUUUGAAGGCAAUGAAGGCCGAUCAUAAAAAGCAGUAAUAAAAAUGGCGGCCAUAUUUUAAAUGGAAUGUACAAUCCCACAUAUUUUAUAAUGCCAGUGCCCGUGAAAAAUGCACCUC .........((((.....(((((.((((.................))))))))).........(((((((((..........)))))))))..)))).((((...(....)..))))... ( -24.53) >DroSec_CAF1 49709 120 + 1 AAAUUUGAAGGCAAUGAAGGCCGAUCAUAAAAAGCAGUAAUAAAAAUGGCGGCCAUAUUUUAAAUGGAAUGUACAAUGCCACAUAUUUUAUAAUGCCAGUGCACGAAAAAAAUGCACCUC .........((((.((..(((((.((((.................)))))))))(((((((....))))))).)).))))..................(((((.........)))))... ( -25.33) >DroSim_CAF1 51012 120 + 1 AAAUUUGAAGGCAAUGAAGGCCAAUCAUAAAAAGCAGUAAUAAAAAUGGCGGCCAUAUUUUAAAUGGAAUGUACAAUGCCACAUAUUUUAUAAUGCCAGUGCACGUAAAAAAUGCACCUC .........((((.((..((((..((((.................)))).))))(((((((....))))))).)).)))).(((.((((((..(((....))).)))))).)))...... ( -21.33) >DroEre_CAF1 42833 120 + 1 AAAUUUGAAGGCAAAUAAGGCCGAUCACAAAAAGCAGUAAUAAUAAUGGCGGCCAUAAUUUAAAUGGAAUGUACAAUCCCACAUAUUUUAUAAUGCCAGCGCACGUAAAAAACGCACCUC .........((((.....(((((.(((...................))))))))((((..((..(((.((.....)).)))..))..))))..)))).(((...........)))..... ( -21.61) >DroYak_CAF1 45108 120 + 1 AAAUUGGAAGGCAAAUAAGAGCGAUCACAAAAAGAGGUUAUACAAAUGGCGGCCAUAUAUUAAAUGGAAUGUACAAUCCAACAUAUUUUAUAAUGCCAGUGCACGUAAAAAAUGCACCUC ...(((((..((........))..)).)))...(((((........(((((.((((.......)))).((((........)))).........)))))..(((.........)))))))) ( -19.40) >consensus AAAUUUGAAGGCAAUGAAGGCCGAUCAUAAAAAGCAGUAAUAAAAAUGGCGGCCAUAUUUUAAAUGGAAUGUACAAUCCCACAUAUUUUAUAAUGCCAGUGCACGUAAAAAAUGCACCUC .........((((.....(((((.((((.................))))))))).........(((((((((..........)))))))))..)))).((((...........))))... (-18.39 = -19.59 + 1.20)


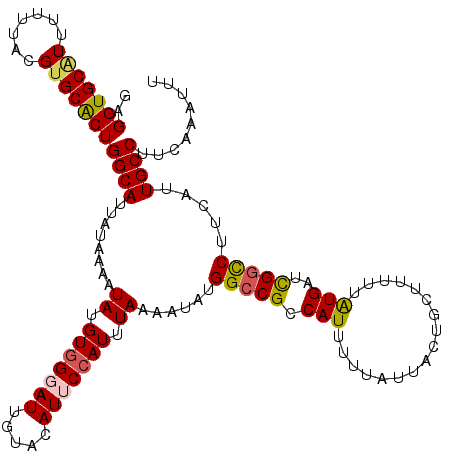
| Location | 20,528,424 – 20,528,544 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.83 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20528424 120 - 22407834 GAGGUGCAUUUUUCACGGGCACUGGCAUUAUAAAAUAUGUGGGAUUGUACAUUCCAUUUAAAAUAUGGCCGCCAUUUUUAUUACUGCUUUUUAUGAUCGGCCUUCAUUGCCUUCAAAUUU ..(((((...........)))))((((........((.(((((((.....))))))).))......(((((.(((.................)))..))))).....))))......... ( -24.53) >DroSec_CAF1 49709 120 - 1 GAGGUGCAUUUUUUUCGUGCACUGGCAUUAUAAAAUAUGUGGCAUUGUACAUUCCAUUUAAAAUAUGGCCGCCAUUUUUAUUACUGCUUUUUAUGAUCGGCCUUCAUUGCCUUCAAAUUU ((((.(((........(((((.((.(((..........))).)).)))))................(((((.(((.................)))..))))).....)))))))...... ( -25.93) >DroSim_CAF1 51012 120 - 1 GAGGUGCAUUUUUUACGUGCACUGGCAUUAUAAAAUAUGUGGCAUUGUACAUUCCAUUUAAAAUAUGGCCGCCAUUUUUAUUACUGCUUUUUAUGAUUGGCCUUCAUUGCCUUCAAAUUU ..(((((((.......)))))))((((..((((((...(((((..(((..............)))..)))))...))))))...)))).....(((..(((.......))).)))..... ( -25.04) >DroEre_CAF1 42833 120 - 1 GAGGUGCGUUUUUUACGUGCGCUGGCAUUAUAAAAUAUGUGGGAUUGUACAUUCCAUUUAAAUUAUGGCCGCCAUUAUUAUUACUGCUUUUUGUGAUCGGCCUUAUUUGCCUUCAAAUUU .(((..(((.....)))..).))((((..((((..((.(((((((.....))))))).))......(((((........(((((........)))))))))))))).))))......... ( -29.10) >DroYak_CAF1 45108 120 - 1 GAGGUGCAUUUUUUACGUGCACUGGCAUUAUAAAAUAUGUUGGAUUGUACAUUCCAUUUAAUAUAUGGCCGCCAUUUGUAUAACCUCUUUUUGUGAUCGCUCUUAUUUGCCUUCCAAUUU (((((((((.......))))..((((........(((((((((((.(......).)))))))))))....))))........)))))...(((.((..((........))..)))))... ( -24.20) >consensus GAGGUGCAUUUUUUACGUGCACUGGCAUUAUAAAAUAUGUGGGAUUGUACAUUCCAUUUAAAAUAUGGCCGCCAUUUUUAUUACUGCUUUUUAUGAUCGGCCUUCAUUGCCUUCAAAUUU ..(((((((.......)))))))((((........((.(((((((.....))))))).))......(((((.(((.................)))..))))).....))))......... (-21.67 = -21.83 + 0.16)

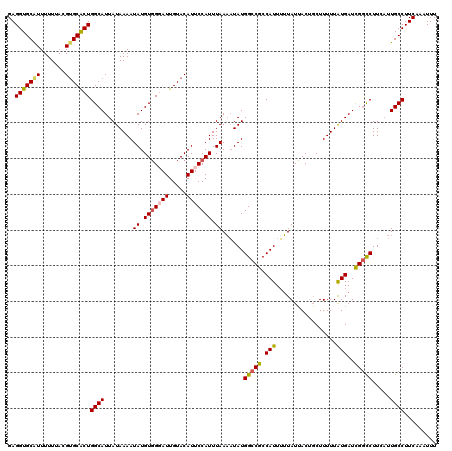
| Location | 20,528,464 – 20,528,583 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.86 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -20.64 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20528464 119 - 22407834 GCGAAAAAAAACACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCAUUUUUCACGGGCACUGGCAUUAUAAAAUAUGUGGGAUUGUACAUUCCAUUUAAAAUAUGGCCGCCAUUUUUA (.((((((.......((((((((((..........))))))))))..)))))).).(((...(((............(((((((.....)))))))..........))))))....... ( -26.15) >DroSec_CAF1 49749 116 - 1 CUGAA---AAACACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCAUUUUUUUCGUGCACUGGCAUUAUAAAAUAUGUGGCAUUGUACAUUCCAUUUAAAAUAUGGCCGCCAUUUUUA ..(((---(((....((((((((((..........))))))))))....))))))(((((.((.(((..........))).)).)))))...((((.......))))............ ( -26.50) >DroSim_CAF1 51052 116 - 1 CUGAA---AAACACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCAUUUUUUACGUGCACUGGCAUUAUAAAAUAUGUGGCAUUGUACAUUCCAUUUAAAAUAUGGCCGCCAUUUUUA .((((---(((....((((((((((..........))))))))))..))))))).(((((.((.(((..........))).)).)))))...((((.......))))............ ( -24.10) >DroEre_CAF1 42873 117 - 1 CUGGAA--AAACACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCGUUUUUUACGUGCGCUGGCAUUAUAAAAUAUGUGGGAUUGUACAUUCCAUUUAAAUUAUGGCCGCCAUUAUUA ...(((--((((...((((((((((..........)))))))))).)))))))..(((.((.(((..(((((..((.(((((((.....))))))).))..)))))))))))))..... ( -29.40) >DroYak_CAF1 45148 116 - 1 CUAAAA---AAAACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCAUUUUUUACGUGCACUGGCAUUAUAAAAUAUGUUGGAUUGUACAUUCCAUUUAAUAUAUGGCCGCCAUUUGUA ....((---((((..((((((((((..........))))))))))..))))))..((((((..((((........))))..)..)))))..........(((.((((...)))).))). ( -26.30) >consensus CUGAAA__AAACACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCAUUUUUUACGUGCACUGGCAUUAUAAAAUAUGUGGGAUUGUACAUUCCAUUUAAAAUAUGGCCGCCAUUUUUA ................(((((((((..........)))))))))((((.......))))..((((.(((((......(((((((.....)))))))......)))))..))))...... (-20.64 = -21.32 + 0.68)



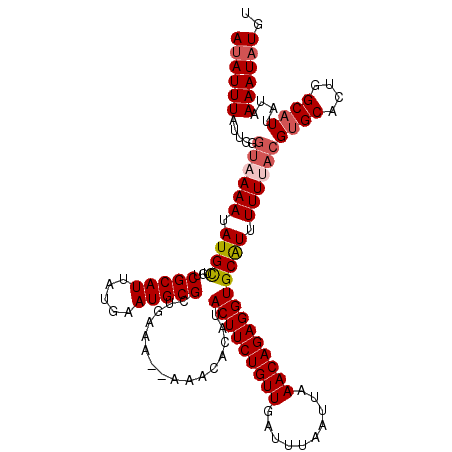
| Location | 20,528,504 – 20,528,623 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.03 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -22.78 |
| Energy contribution | -23.66 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.978969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20528504 119 - 22407834 AUAUUUAUUGGGUAAAAUAUGUUGUCGCAUUAUGAAUGCGGCGAAAAAAAACACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCAUUUUUCACGGGCACUGGCAUUAUAAAAUAUGU ((((((.....((.(((.((((((((((((.....)))))))))...........((((((((((..........))))))))))))).))).))..((....))......)))))).. ( -25.90) >DroSec_CAF1 49789 116 - 1 AUAUUUAUUGGGUAAAAUAUGCUGUCGCAUUAUGAAUGCGCUGAA---AAACACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCAUUUUUUUCGUGCACUGGCAUUAUAAAAUAUGU .................((((.((((((((.....))))((((((---(((....((((((((((..........))))))))))....))))))).))...)))).))))........ ( -26.70) >DroSim_CAF1 51092 116 - 1 AUAUUUAUUGGGUAAAAUAUGCUGUCGCAUUAUGAAUGCGCUGAA---AAACACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCAUUUUUUACGUGCACUGGCAUUAUAAAAUAUGU ((((((.....((((((.((((.(.(((((.....))))))....---........(((((((((..........))))))))))))).))))))((((....))))....)))))).. ( -28.00) >DroEre_CAF1 42913 117 - 1 AUAUUUAUUGGUUCAAAUAUGCUGUCGCAUUAUGAAUGCGCUGGAA--AAACACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCGUUUUUUACGUGCGCUGGCAUUAUAAAAUAUGU ((((((....(((((...((((....))))..)))))((((..(((--((((...((((((((((..........)))))))))).)))))))....))))..........)))))).. ( -30.90) >DroYak_CAF1 45188 116 - 1 AAAUUUAUCGGGUAAAAUAUGCCGUCGCAUUAUGAAUGCGCUAAAA---AAAACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCAUUUUUUACGUGCACUGGCAUUAUAAAAUAUGU ...........((((((.((((.(.(((((.....)))))).....---.......(((((((((..........))))))))))))).))))))((((....))))............ ( -26.50) >consensus AUAUUUAUUGGGUAAAAUAUGCUGUCGCAUUAUGAAUGCGCUGAAA__AAACACAUACUUCUGUUGAUUUAAUUAAACAGAGGUGCAUUUUUUACGUGCACUGGCAUUAUAAAAUAUGU ((((((.....((((((.((((...(((((.....)))))................(((((((((..........))))))))))))).))))))((((....))))....)))))).. (-22.78 = -23.66 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:06 2006