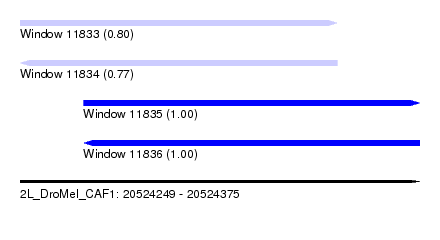
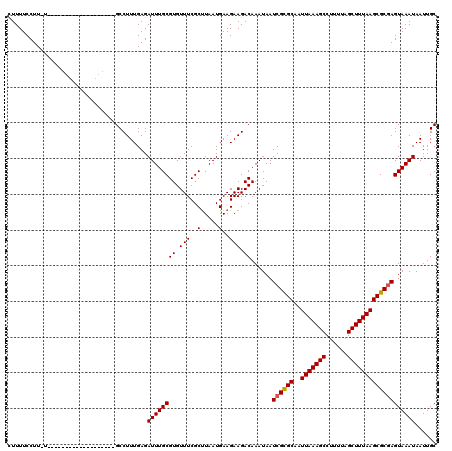
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,524,249 – 20,524,375 |
| Length | 126 |
| Max. P | 0.999992 |

| Location | 20,524,249 – 20,524,349 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.17 |
| Mean single sequence MFE | -24.57 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20524249 100 + 22407834 UUUUUCCUU-U-------------------GCCUUUGAGAUUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGC .......((-(-------------------(.((((...(((.(((.....)))..)))...)))).))))...((((((..(((((((......)))))))))))))............ ( -23.80) >DroSim_CAF1 46618 100 + 1 CUUUUCCUU-U-------------------GCCUUUGAGAUUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGC .......((-(-------------------(.((((...(((.(((.....)))..)))...)))).))))...((((((..(((((((......)))))))))))))............ ( -23.80) >DroEre_CAF1 38740 112 + 1 CUUUUCCUU-UU-------CCUUUGCCUUCGCCUUUGAGAUUUGCGUGUUUCGCCUAAUGAAGAAGACAAAUAAUCGUGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGC .........-..-------..((((((((((....((((((......)))))).....)))))...........((((((..(((((((......))))))))))))))))))....... ( -22.80) >DroYak_CAF1 40903 100 + 1 CUUUUCCUU-U-------------------GCCUUUGAGAUUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGC .......((-(-------------------(.((((...(((.(((.....)))..)))...)))).))))...((((((..(((((((......)))))))))))))............ ( -23.80) >DroAna_CAF1 38493 101 + 1 CUUUUCCUUUU-------------------GCCUUUGAGAUUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGC ........(((-------------------(.((((...(((.(((.....)))..)))...)))).))))...((((((..(((((((......)))))))))))))............ ( -23.90) >DroPer_CAF1 88185 119 + 1 CUUUUCCUU-GUGCUGGUGCCUUUGCCUUUGCCUUUGAGAUUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCAAGUAAAUAAUUGC ..(((.(((-(((((.((((..(((..((((.((((...(((.(((.....)))..)))...)))).)))))))..))))...((((((......)))))))))))))).)))....... ( -29.30) >consensus CUUUUCCUU_U___________________GCCUUUGAGAUUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGC .......................................((((((((.(((..(.....)..))).))......((((((..(((((((......)))))))))))))))))))...... (-22.25 = -22.28 + 0.03)

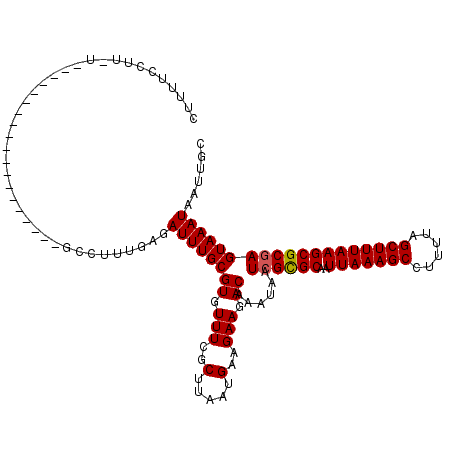
| Location | 20,524,249 – 20,524,349 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.17 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -20.83 |
| Energy contribution | -20.86 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20524249 100 - 22407834 GCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGC-------------------A-AAGGAAAAA ............((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........)))))-------------------)-))....... ( -25.93) >DroSim_CAF1 46618 100 - 1 GCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGC-------------------A-AAGGAAAAG ............((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........)))))-------------------)-))....... ( -25.93) >DroEre_CAF1 38740 112 - 1 GCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCACGAUUAUUUGUCUUCUUCAUUAGGCGAAACACGCAAAUCUCAAAGGCGAAGGCAAAGG-------AA-AAGGAAAAG ((..........(((.((((((((((....))))))))..)).)))...(((((((........)))))))....)).....((....((....))....)-------).-......... ( -24.60) >DroYak_CAF1 40903 100 - 1 GCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGC-------------------A-AAGGAAAAG ............((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........)))))-------------------)-))....... ( -25.93) >DroAna_CAF1 38493 101 - 1 GCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGC-------------------AAAAGGAAAAG ............((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........)))))-------------------)))........ ( -25.93) >DroPer_CAF1 88185 119 - 1 GCAAUUAUUUACUUGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGCAAAGGCAAAGGCACCAGCAC-AAGGAAAAG ((..........((((((((((((((....))))))))..))))))...((((((((.........(((.....)))........)))))))).))....((....))..-......... ( -27.03) >consensus GCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAAUCUCAAAGGC___________________A_AAGGAAAAG ............((((((((((((((....))))))))..))))))......(((((.........(((.....)))........))))).............................. (-20.83 = -20.86 + 0.03)

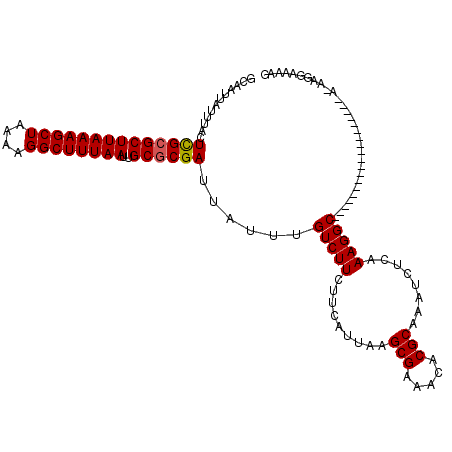

| Location | 20,524,269 – 20,524,375 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -31.97 |
| Energy contribution | -31.75 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.67 |
| SVM RNA-class probability | 0.999992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20524269 106 + 22407834 UUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAAACGAACGCACGCA ..(((((((((((.....................((((((..(((((((......)))))))))))))...........((((....))))...)))).))))))) ( -32.70) >DroSec_CAF1 45696 106 + 1 UUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAAACGAACGCACGCA ..(((((((((((.....................((((((..(((((((......)))))))))))))...........((((....))))...)))).))))))) ( -32.70) >DroSim_CAF1 46638 106 + 1 UUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAAACGAACGCACGCA ..(((((((((((.....................((((((..(((((((......)))))))))))))...........((((....))))...)))).))))))) ( -32.70) >DroYak_CAF1 40923 105 + 1 UUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAA-CGAACACACGCA ..(((((((((((.....................((((((..(((((((......)))))))))))))...........((((....))))..-)))).))))))) ( -32.70) >DroAna_CAF1 38514 106 + 1 UUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAAACGAACACACGCA ..(((((((((((.....................((((((..(((((((......)))))))))))))...........((((....))))...)))).))))))) ( -32.70) >DroPer_CAF1 88224 105 + 1 UUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCAAGUAAAUAAUUGCUGUGAGUGCAGAA-CGAGAACACGCA ..(((((((((((((((...................((((..(((((((......))))))))))).(((((....))))).)))))).....-...))))))))) ( -30.00) >consensus UUUGCGUGUUUCGCUUAAUGAAGAAGACAAAUAAUCGCGCAAUUAAAGCCUUUUAGCUUUAAGCGCGAGUAAAUAAUUGCUGUGAGUGCAGAAACGAACACACGCA ..(((((((((((.....................((((((..(((((((......)))))))))))))...........((((....))))...)))).))))))) (-31.97 = -31.75 + -0.22)

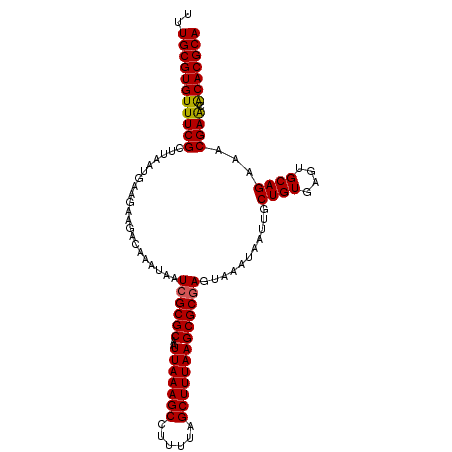
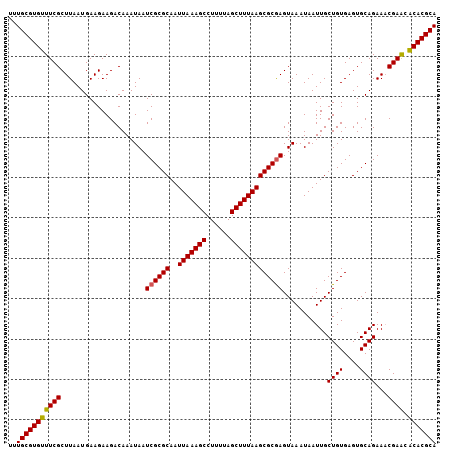
| Location | 20,524,269 – 20,524,375 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -30.92 |
| Energy contribution | -31.12 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.72 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20524269 106 - 22407834 UGCGUGCGUUCGUUUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAA ((((((..(((((((.(((.......)))..((..((.((((((((((((((....))))))))..)))))).))..))...........))))))).)))))).. ( -34.00) >DroSec_CAF1 45696 106 - 1 UGCGUGCGUUCGUUUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAA ((((((..(((((((.(((.......)))..((..((.((((((((((((((....))))))))..)))))).))..))...........))))))).)))))).. ( -34.00) >DroSim_CAF1 46638 106 - 1 UGCGUGCGUUCGUUUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAA ((((((..(((((((.(((.......)))..((..((.((((((((((((((....))))))))..)))))).))..))...........))))))).)))))).. ( -34.00) >DroYak_CAF1 40923 105 - 1 UGCGUGUGUUCG-UUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAA (((((((.((((-((.(((.......)))..((..((.((((((((((((((....))))))))..)))))).))..))............))))))))))))).. ( -34.90) >DroAna_CAF1 38514 106 - 1 UGCGUGUGUUCGUUUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAA (((((((.(((((((.(((.......)))..((..((.((((((((((((((....))))))))..)))))).))..))...........)))))))))))))).. ( -35.70) >DroPer_CAF1 88224 105 - 1 UGCGUGUUCUCG-UUCUGCACUCACAGCAAUUAUUUACUUGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAA ((((((((.(((-((.(((.......)))..((..((.((((((((((((((....))))))))..)))))).))..))............))))))))))))).. ( -32.80) >consensus UGCGUGCGUUCGUUUCUGCACUCACAGCAAUUAUUUACUCGCGCUUAAAGCUAAAAGGCUUUAAUUGCGCGAUUAUUUGUCUUCUUCAUUAAGCGAAACACGCAAA ((((((...((((((.(((.......)))..((..((.((((((((((((((....))))))))..)))))).))..))...........))))))..)))))).. (-30.92 = -31.12 + 0.19)

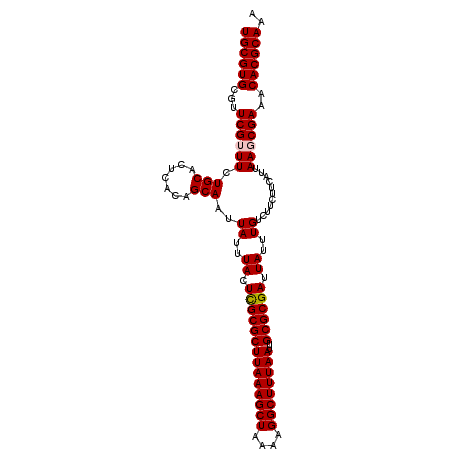
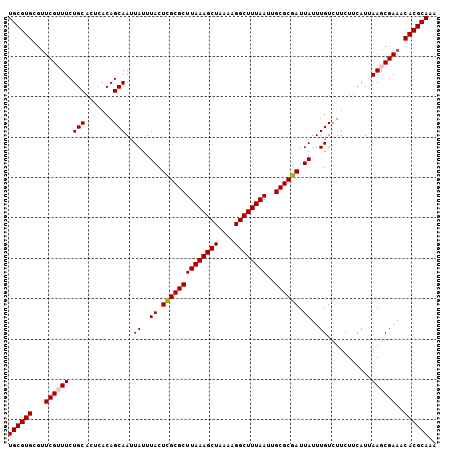
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:35:01 2006