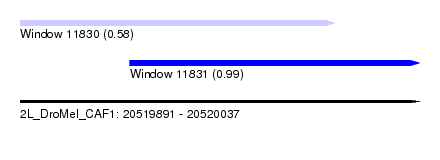
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,519,891 – 20,520,037 |
| Length | 146 |
| Max. P | 0.987381 |

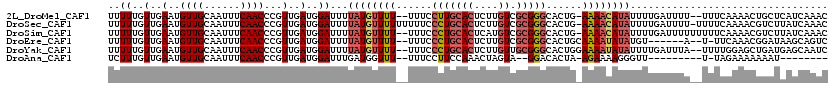
| Location | 20,519,891 – 20,520,006 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -13.38 |
| Energy contribution | -14.18 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20519891 115 + 22407834 UUUUUGUUGAAUGUUGCAAUUUCAACCCGUUGAUGGAUUUUAUGUUUU--UUUCCUUGCACUCUUGUCGCGGGCACUG-AAAACAUAUUUUGAUUUU--UUUCAAAACUGCUCAUCAAAC .....((((((.........))))))...(((((((....((((((((--(...(((((((....)).)))))....)-))))))))((((((....--..))))))....))))))).. ( -26.20) >DroSec_CAF1 41222 118 + 1 UUUUUGUUGAAUGUUGCAAUUUCAACCCGUUGAUGGAUUUUAUGUUUUUUUUUCCCUGCACUCUUGUCGCGGGCACUG-AAAACAUAUUUUGAUUUU-UUUUCAAAACGUCUUAUCAAAC .....((((((.........))))))...(((((((((..(((((((((.....(((((((....)).)))))....)-))))))))((((((....-...)))))).))).)))))).. ( -27.30) >DroSim_CAF1 42088 117 + 1 UUUUUGUUGAAUGUUGCAAUUUCAACCCGUUGAUGGAUUUUAUGUUUU--UUUCCCUGCACUCAUGUCGCGGGCACUG-AAAACAUAUUUUGAUUUUUUUUUCAAAACGUCUUAUCAAAC .....((((((.........))))))...(((((((((..((((((((--(...(((((((....)).)))))....)-))))))))((((((........)))))).))).)))))).. ( -27.80) >DroEre_CAF1 34775 109 + 1 UUUUUGUUGAAUGUUGCAAUUUCAACCCGUUGAUGGAUUUUAUGUUUU--UUUCCCUGCACUCUUGUCGCGGGCACUGCAAAAUAUAUGU------A--U-UUCAAACGGAUAAGCAGUC .....((((((.........))))))(((((...((((..((((((((--....(((((((....)).)))))......))))))))...------)--)-))..))))).......... ( -22.60) >DroYak_CAF1 36433 116 + 1 UUUUUGUUGAAUGUUGCAAUUUCAACCCGUUGAUGGAUUUUAUGUUUU--UUUCCCUGCACUCUUGUUGCGGGCACUGGAAAAUAUAUUUUGAUUUA--UUUUGGAGCUGAUGAGCAAUC .....((((((.........))))))(((..((((((((.((((((((--((..(((((((....).))))))....))))))))))....))))))--)).))).(((....))).... ( -27.60) >DroAna_CAF1 32932 97 + 1 UCUUUGUUGAAUGUUGCAAUUUCAACCCGUUGAUGGAUUUGAUGGUUU--UUUCCUUCCAAACUAGUA--GGACACUA-AGAAAAGGGUU---------U-UAGAAAAAAAU-------- .....((((((.........))))))((((..(.....)..))))(((--((((...((.......((--(....)))-.......))..---------.-..)))))))..-------- ( -16.34) >consensus UUUUUGUUGAAUGUUGCAAUUUCAACCCGUUGAUGGAUUUUAUGUUUU__UUUCCCUGCACUCUUGUCGCGGGCACUG_AAAACAUAUUUUGAUUUU__UUUCAAAACGGAUUAUCAAAC ..((..(..(..((((......))))...)..)..))...((((((((......(((((((....)).)))))......))))))))................................. (-13.38 = -14.18 + 0.81)


| Location | 20,519,931 – 20,520,037 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.86 |
| Mean single sequence MFE | -17.74 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20519931 106 + 22407834 UAUGUUUU--UUUCCUUGCACUCUUGUCGCGGGCACUG-AAAACAUAUUUUGAUUUU--UUUCAAAACUGCUCAUCAAACACACCCCUAUACA----AAUUU-----AAGAUAUCUAUGA ((((((((--(...(((((((....)).)))))....)-))))))))((((((....--..))))))..........................----.....-----............. ( -15.60) >DroSec_CAF1 41262 108 + 1 UAUGUUUUUUUUUCCCUGCACUCUUGUCGCGGGCACUG-AAAACAUAUUUUGAUUUU-UUUUCAAAACGUCUUAUCAAACAUCCCCCUAUACA----CAUUU-----ACGAUAUCUAUG- (((((((((.....(((((((....)).)))))....)-))))))))((((((....-...))))))..........................----.....-----............- ( -17.70) >DroSim_CAF1 42128 107 + 1 UAUGUUUU--UUUCCCUGCACUCAUGUCGCGGGCACUG-AAAACAUAUUUUGAUUUUUUUUUCAAAACGUCUUAUCAAACAUCCCCCUAUACA----CAUUU-----ACGAUAUCUAUG- ((((((((--(...(((((((....)).)))))....)-))))))))((((((........))))))..........................----.....-----............- ( -18.20) >DroEre_CAF1 34815 109 + 1 UAUGUUUU--UUUCCCUGCACUCUUGUCGCGGGCACUGCAAAAUAUAUGU------A--U-UUCAAACGGAUAAGCAGUCAUUUAAUUAUAUAUUUAAAUCUAUCGAAAAAUAUCUAUCU .(((((((--((..(((((((....)).))))).(((((....(((.(((------.--.-.....))).))).))))).((((((........)))))).....)))))))))...... ( -16.00) >DroYak_CAF1 36473 98 + 1 UAUGUUUU--UUUCCCUGCACUCUUGUUGCGGGCACUGGAAAAUAUAUUUUGAUUUA--UUUUGGAGCUGAUGAGCAAUCAUAUAAUUUU------------------AAAUAUCUAUCU ((((((((--((..(((((((....).))))))....))))))))))....(((.((--(((..((....((((....))))....))..------------------)))))...))). ( -21.20) >consensus UAUGUUUU__UUUCCCUGCACUCUUGUCGCGGGCACUG_AAAACAUAUUUUGAUUUU__UUUCAAAACGGCUUAUCAAACAUACCCCUAUACA____AAUUU_____AAGAUAUCUAUG_ ((((((((......(((((((....)).)))))......))))))))((((((........))))))..................................................... (-13.32 = -13.16 + -0.16)

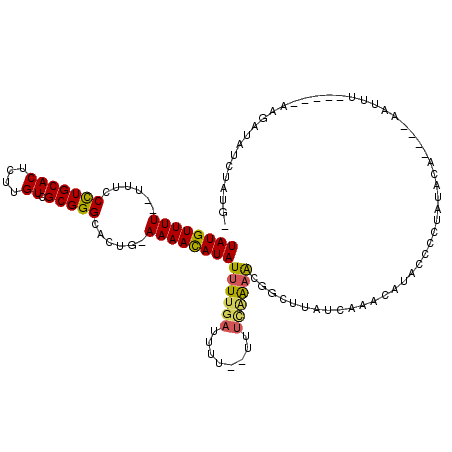
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:56 2006