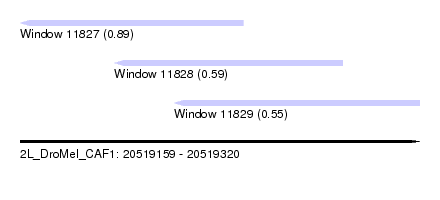
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,519,159 – 20,519,320 |
| Length | 161 |
| Max. P | 0.890886 |

| Location | 20,519,159 – 20,519,249 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -14.75 |
| Consensus MFE | -10.95 |
| Energy contribution | -11.12 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
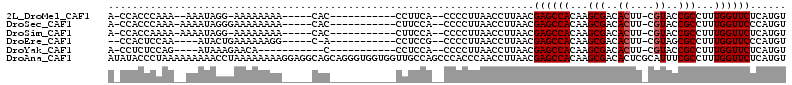
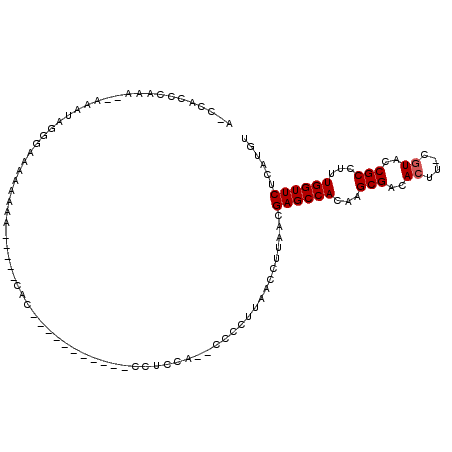
>2L_DroMel_CAF1 20519159 90 - 22407834 A-CCACCCAAA--AAAUAGG-AAAAAAAA-----CAC-----------CCUUCA--CCCCUUAACCUUAACGAGCCACAAGCGACACUU-CGUACCGCCUUUGGUUCUCAUGU .-....((...--.....))-........-----...-----------......--...............((((((...(((......-.....)))...))))))...... ( -11.00) >DroSec_CAF1 40474 92 - 1 A-CCACCCAAA-AAAAUAGGGAAAAAAAA-----CAC-----------CUUCCA--CCCCUUAACCUUAACGAGCCACAAGCGACACUU-CGUACCGCCUUUGGUUCCCAUGU .-...(((...-......)))........-----...-----------......--...............((((((...(((......-.....)))...))))))...... ( -13.30) >DroSim_CAF1 41345 91 - 1 A-CCACCAAAA-AAAAUAGG-AAAAAAAA-----CAC-----------CUUCCA--CCCCUUAACCUUAACGAGCCACAAGCGACACUU-CGUACCGCCUUUGGUUCUCAUGU .-.........-......((-((......-----...-----------.)))).--...............((((((...(((......-.....)))...))))))...... ( -12.90) >DroEre_CAF1 34143 87 - 1 --CCACUCCAA----AUACUGAAAAAAGG-----C-A-----------CCUCCG--CCCCUUAACCUUAACGAGCCACAAGCGACACUU-CGUAGCGCCUUUGGUUCCCAUGU --.........----............((-----(-.-----------.....)--)).............((((((...(((..((..-.))..)))...))))))...... ( -14.70) >DroYak_CAF1 35747 83 - 1 A-CCUCUCCAG----AUAAAGAACA-----------C-----------CCUCCA--CCCCUUAACCUUAACGAGCCACAAGCGACACUU-CGUACCGCCUUUGGUUCUCAUGU .-.........----..........-----------.-----------......--...............((((((...(((......-.....)))...))))))...... ( -10.40) >DroAna_CAF1 32135 113 - 1 AUAUACCCUAAAAAAAAACCUAAAAAAAAGGAGGCAGCAGGGUGGUGGUUGCCAGCCCACCCAACCUUAACGAGCCACAAGCGACACUCGCAUUUCGCCUUUGGUUCUCAUGU ..................(((.......)))..(((...(((((((((...)))..)))))).........((((((...((((..........))))...))))))...))) ( -26.20) >consensus A_CCACCCAAA__AAAUAGGGAAAAAAAA_____CAC___________CCUCCA__CCCCUUAACCUUAACGAGCCACAAGCGACACUU_CGUACCGCCUUUGGUUCUCAUGU .......................................................................((((((...(((..((....))..)))...))))))...... (-10.95 = -11.12 + 0.17)
| Location | 20,519,197 – 20,519,289 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -13.92 |
| Consensus MFE | -6.52 |
| Energy contribution | -6.20 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20519197 92 - 22407834 CACUCCACGGUUAUCAGUGGCAGCCCAACAGCUAGUCCGCACCACCCAAA-AAAUAGG-AAAAAAAACACCCUUCACCCCUUAACCUUAACGAG ..(((...(((((...((((((((......))).).)))).....((...-.....))-......................))))).....))) ( -13.80) >DroSec_CAF1 40512 94 - 1 CACUCCUCGGUUAUCAGUGGCAGCCCAACAGCUAGCCCUCACCACCCAAAAAAAUAGGGAAAAAAAACACCUUCCACCCCUUAACCUUAACGAG .....((((((....((.((((((......))).))))).))).(((.........)))................................))) ( -16.00) >DroSim_CAF1 41383 93 - 1 CACUCCUCGGUUAUCAGUGGCAGCCCAACAGCUAGCCCUCACCACCAAAAAAAAUAGG-AAAAAAAACACCUUCCACCCCUUAACCUUAACGAG .....((((((....((.((((((......))).)))))....)))..........((-((..........))))................))) ( -16.20) >DroEre_CAF1 34181 77 - 1 CACUCCUCGGUUAUCAGUGGCAGCCCAA-------------CCACUCCAA---AUACUGAAAAAAGGC-ACCUCCGCCCCUUAACCUUAACGAG ..(((.(((((....(((((........-------------)))))....---..))))).....(((-......))).............))) ( -14.80) >DroYak_CAF1 35785 85 - 1 CACUCCUCGGUUAUCAGUGGUAUUCCAACAGCUAGCCCACACCUCUCCAG---AUAAAGAACA------CCCUCCACCCCUUAACCUUAACGAG .....((((((((.(..(((....)))...).)))))......(((....---....)))...------......................))) ( -8.80) >consensus CACUCCUCGGUUAUCAGUGGCAGCCCAACAGCUAGCCCUCACCACCCAAA_AAAUAGGGAAAAAAAACACCCUCCACCCCUUAACCUUAACGAG ..(((...(((((...((((.....................................................))))....))))).....))) ( -6.52 = -6.20 + -0.32)

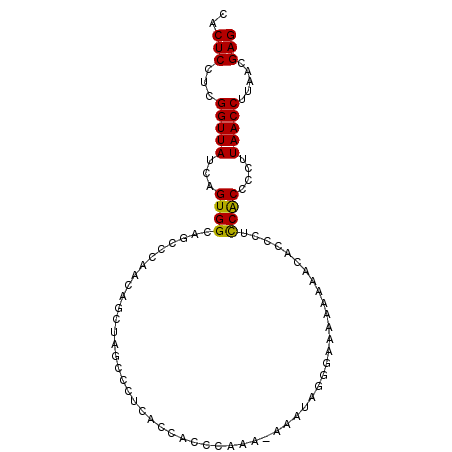

| Location | 20,519,221 – 20,519,320 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.23 |
| Mean single sequence MFE | -16.73 |
| Consensus MFE | -8.30 |
| Energy contribution | -9.18 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20519221 99 - 22407834 UCACGCUGCAAAUGAUAUCCAAAGACUAAUCCACUCCACGGUUAUCAGUGGCAGCCCAACAGCUAGUCCGCA-CCACCCAAA--AAAUAGG-AAAAAAAA-----CAC--- ....(((((...(((((.((...................)).)))))...)))))......((......)).-....((...--.....))-........-----...--- ( -17.01) >DroSec_CAF1 40536 101 - 1 ACACGCUGCAAAUGAUAUCCAAAGACUAAUCCACUCCUCGGUUAUCAGUGGCAGCCCAACAGCUAGCCCUCA-CCACCCAAA-AAAAUAGGGAAAAAAAA-----CAC--- ....(((((...(((((.((...................)).)))))...))))).................-...(((...-......)))........-----...--- ( -18.41) >DroSim_CAF1 41407 100 - 1 ACACGCUGCAAAUGAUAUCCAAAGACUAAUCCACUCCUCGGUUAUCAGUGGCAGCCCAACAGCUAGCCCUCA-CCACCAAAA-AAAAUAGG-AAAAAAAA-----CAC--- ....(((((...(((((.((...................)).)))))...))))).................-.........-........-........-----...--- ( -15.11) >DroYak_CAF1 35809 92 - 1 ACCCGCUGCAAACGAUAACCAGAGACUAAUCCACUCCUCGGUUAUCAGUGGUAUUCCAACAGCUAGCCCACA-CCUCUCCAG----AUAAAGAACA-----------C--- ....(((((....(((((((.(((............))))))))))..(((....)))...)).))).....-.........----..........-----------.--- ( -18.50) >DroAna_CAF1 32208 100 - 1 -UACAUGGCAAAGGACUUGCUGAGACUAAUCCACUCUCCGGUUAUCAGUGGUACUUC----------CCUUAUAUACCCUAAAAAAAAACCUAAAAAAAAGGAGGCAGCAG -................(((((...((............((((((.((.((.....)----------))).))).)))...........(((.......))))).))))). ( -14.60) >consensus ACACGCUGCAAAUGAUAUCCAAAGACUAAUCCACUCCUCGGUUAUCAGUGGCAGCCCAACAGCUAGCCCUCA_CCACCCAAA_AAAAUAGG_AAAAAAAA_____CAC___ ....(((((...(((((.((.(.((.........)).).)).)))))...)))))........................................................ ( -8.30 = -9.18 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:55 2006