
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,515,820 – 20,515,998 |
| Length | 178 |
| Max. P | 0.949917 |

| Location | 20,515,820 – 20,515,918 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.74 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -8.92 |
| Energy contribution | -9.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20515820 98 - 22407834 AAUUGCCUGGAAAGCACAUGGGUGGAUA--------------UCCUGCUUGAAUAUUGUUUGUGUGGG---GCUGCUCAUUUUG---UAGUCCAAAGGAUAUCGCCCACACA--AAGCGA ..((((((....))....((((((.(((--------------((((......((((.....)))).((---(((((.......)---))))))..)))))))))))))....--..)))) ( -31.30) >DroSec_CAF1 37201 98 - 1 AAUUGCCUGGAAAGCACAUGGGUGGAUA--------------UCCUGCUCGAAUAUUGUUUGUGUGGG---GCUGCUCAUUUUG---UAGUCCAAAGGAUAUCGCCCACACA--AAGCGA ..((((((....))....((((((.(((--------------((((((.(((((...))))).)).((---(((((.......)---))))))..)))))))))))))....--..)))) ( -33.80) >DroSim_CAF1 38013 98 - 1 AAUUGCCUGGAAAGCACAUGGGUGGAUA--------------UCCUGCUCGAAUAUUGUUUGUGUGGG---GCUGCUCAUUUUG---UAGUCCAAAGGAUAUCGUUCACACA--AAGCGA ..(((..((((...(((....)))((((--------------((((((.(((((...))))).)).((---(((((.......)---))))))..)))))))).))))..))--)..... ( -28.40) >DroYak_CAF1 32426 101 - 1 AAUUGCCUGGAAAUCACAAGGGUAGAUA--------------UCCUGCUCGAAUAUUGUUUGUGUGGGGGCGCUGCUCAAUUUG---UAGCCCAAAGGAUAUCGCACACACA--AAGCGA ...(((((.(....)....)))))((((--------------((((((((..((((.....))))..))))(((((.......)---))))....))))))))((.......--..)).. ( -29.30) >DroAna_CAF1 25028 115 - 1 AAUUGCCUGGAAAGCACAUGGGUGGAUACGUCCCUACAUGUAUCCUGCCCCA-CAUUGU----CUGGAAACAUUUUUGCCUUUGUGAUAUCCGCAUGGAUAUCGCACACACACUGAGAAA ...(((.(....))))...((((((((((((......)))))))).))))..-((.(((----.((....))..........((((((((((....))))))))))...))).))..... ( -33.60) >consensus AAUUGCCUGGAAAGCACAUGGGUGGAUA______________UCCUGCUCGAAUAUUGUUUGUGUGGG___GCUGCUCAUUUUG___UAGUCCAAAGGAUAUCGCACACACA__AAGCGA ..((((.............((((((...................))))))...........(((((((....((......................))......))))))).....)))) ( -8.92 = -9.32 + 0.40)

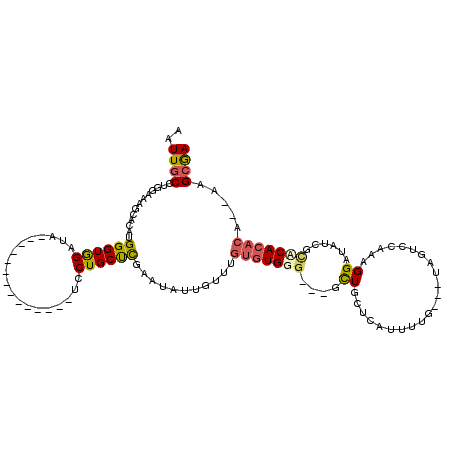
| Location | 20,515,890 – 20,515,998 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -13.55 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20515890 108 - 22407834 GCGCGACUUCCCCACCUUUCUGUUGUCUCGUGUUUAUUGAAAUUUCGUUUUACGACUUUCAAUAUAAAAUUUAAUUAGGCAAUUGCCUGGAAAGCACAUGGGUGGAUA------------ ...........((((((...((((.((.......((((((((..(((.....))).))))))))...........(((((....))))))).))))...))))))...------------ ( -28.00) >DroPse_CAF1 72013 91 - 1 ------CUUCCCUGGCCUUCU--------GUGCUUAUUGAAAUUUCCUUUUACGACAUUCGAUAUAAAAUUUAAUUAGGCAAUUGCCUGGAAAGCACAUGGGU---UA------------ ------......((((((..(--------((((((((((((...((.......))..))))))...........((((((....)))))).))))))).))))---))------------ ( -22.80) >DroEre_CAF1 30924 101 - 1 G-----CUUCGCCACCUUUUG-CUGUCUCGUGUUUAUUGAGAUCUCGUUUUACGACUUUCGGUGUAAAAUUUAAUUAGGCAAUUGCCUG-ACAGCACACGGGUGGAUG------------ .-----.....((((((..((-(((((.......((((((((..(((.....))).))))))))............((((....)))))-))))))...))))))...------------ ( -33.50) >DroYak_CAF1 32499 108 - 1 CCCCCACUUCACCACCUUUUGGUUGUCUCAUGUUUAUUGAAAUUUCGUUUUACGACUUUCAAUAUAAAAUUUAAUUAGGCAAUUGCCUGGAAAUCACAAGGGUAGAUA------------ .............(((((((((((..........((((((((..(((.....))).))))))))..........((((((....)))))).))))).)))))).....------------ ( -22.90) >DroAna_CAF1 25103 120 - 1 UCCGCAUUUUACCCACUUUGUGUUGUCUUUUGCUUAUUGAAAUUUGGUUUUACGACAUUCGAUAUAAAAUUUAAUUAGGCAAUUGCCUGGAAAGCACAUGGGUGGAUACGUCCCUACAUG ..((...((((((((...((((((.((.......(((((((..(((......)))..)))))))...........(((((....))))))).))))))))))))))..)).......... ( -29.10) >DroPer_CAF1 78135 91 - 1 ------CUUCCCUGGCUUUCU--------GUGCUUAUUGAAAUUUCCUUUUACGACAUUCGAUAUAAAAUUUAAUUAGGCAAUUGCCUCGAAAGCACAUGGGU---UA------------ ------...(((((((((((.--------.....(((((((...((.......))..)))))))............((((....)))).)))))).)).))).---..------------ ( -21.00) >consensus ______CUUCCCCACCUUUCU_UUGUCUCGUGCUUAUUGAAAUUUCGUUUUACGACAUUCGAUAUAAAAUUUAAUUAGGCAAUUGCCUGGAAAGCACAUGGGUGGAUA____________ .............................((((((((((((...(((.....)))..)))))))..........((((((....))))))..)))))....................... (-13.55 = -14.08 + 0.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:52 2006