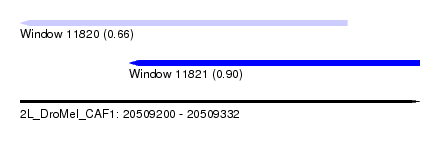
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,509,200 – 20,509,332 |
| Length | 132 |
| Max. P | 0.903537 |

| Location | 20,509,200 – 20,509,308 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -10.87 |
| Energy contribution | -10.32 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
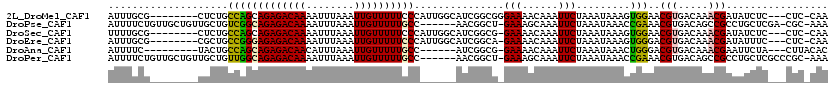
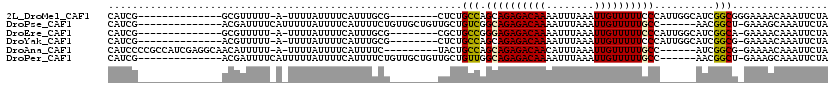
>2L_DroMel_CAF1 20509200 108 - 22407834 AUUUGCG--------CUCUGCCAGCAGAGACAAAAUUUAAAUUGUUUUUCCCAUUGGCAUCGGCGGGAAAACAAAUUCUAAAUAAAGUGGAACGUGACAAACGAUAUCUC---CUC-CAA .....((--------((.((((((.((((((((........))))))))....))))))..))))(((.......(((((.......)))))(((.....))).......---.))-).. ( -22.80) >DroPse_CAF1 65504 111 - 1 AUUUUCUGUUGCUGUUGCUGUCGGCAGAGACAAAAUUUAAAUUGUUUUUGCC------AACGGCU-GAAAGCAAAUUCUAAAUAAACCGAAACGUGACAGCCGCCUGCUCGA-CGC-AAA ((((....(((((...(((((.(((((((((((........)))))))))))------.))))).-...))))).....))))..........(((.((((.....))).).-)))-... ( -32.10) >DroSec_CAF1 26270 107 - 1 UUUUGCG--------CUCUGCCAGCAGAGACAAAAUUUAAAUUGUUUUUCCCAUUGGCAUCGGCG-GAAAACAAAUUCUAAAUAAAGUGGAACGUGACAAACGAUAUCUC---CUC-CAA ((((.((--------((.((((((.((((((((........))))))))....))))))..))))-.))))((..(((((.......)))))..))..............---...-... ( -21.90) >DroEre_CAF1 24904 107 - 1 AUUUGCG--------CGCUGCCGGGAGAGACAAAAUUUAAAUUGUUUUUCCCAUUGGCAUCGGCA-GAAAACAAAUUCUAAAUAAAGUGGGACGUGACAAACGAUAUUUC---CUC-CAA .(((((.--------((.(((((((((((((((........)))))))))))...)))).)))))-))..................(..((((((.....))).....))---)..-).. ( -32.20) >DroAna_CAF1 19040 101 - 1 AUUUUC---------UACUGCCAGCAGAGACAACAUUUAAAUUGUUUUUGCC------AUCGGCG-GAAAACAAAUUCUAAAUAAACUGGGACGUGACAAACGAAUUCUA---CUUACAC ...(((---------..(((((.((((((((((........)))))))))).------...))))-)....((..(((((.......)))))..))......))).....---....... ( -20.90) >DroPer_CAF1 71253 112 - 1 AUUUUCUGUUGCUGUUGCUGUUGGCAGAGACAAAAUUUAAAUUGUUUUUGCC------AACGGCU-GAAAGCAAAUUCUAAAUAAACCGAAACGUGACAGCCGCCUGCUCGCCCGC-AAA ((((....(((((...(((((((((((((((((........)))))))))))------)))))).-...))))).....))))..........(((((((....))).))))....-... ( -37.20) >consensus AUUUGCG________CGCUGCCAGCAGAGACAAAAUUUAAAUUGUUUUUCCC______AUCGGCG_GAAAACAAAUUCUAAAUAAACUGGAACGUGACAAACGAUAUCUC___CUC_CAA ....................(((((((((((((........))))))))))...............(((......))).........)))..(((.....)))................. (-10.87 = -10.32 + -0.55)
| Location | 20,509,236 – 20,509,332 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -9.01 |
| Energy contribution | -8.82 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
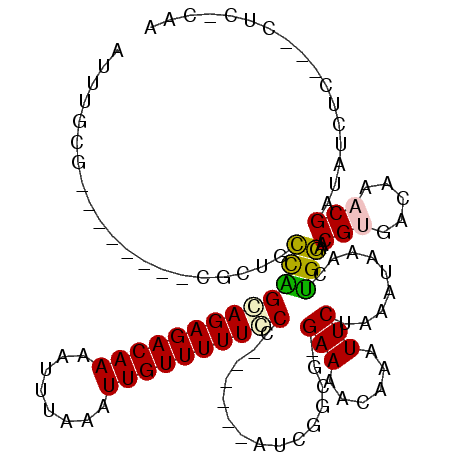
>2L_DroMel_CAF1 20509236 96 - 22407834 CAUCG--------------GCGUUUUU-A-UUUUAUUUUCAUUUGCG--------CUCUGCCAGCAGAGACAAAAUUUAAAUUGUUUUUCCCAUUGGCAUCGGCGGGAAAACAAAUUCUA .....--------------........-.-...........(((.((--------((.((((((.((((((((........))))))))....))))))..)))).)))........... ( -18.80) >DroPse_CAF1 65542 99 - 1 CAUCG--------------ACGAUUUUCAUUUUUAUUUUCAUUUUCUGUUGCUGUUGCUGUCGGCAGAGACAAAAUUUAAAUUGUUUUUGCC------AACGGCU-GAAAGCAAAUUCUA .....--------------.............................(((((...(((((.(((((((((((........)))))))))))------.))))).-...)))))...... ( -27.90) >DroEre_CAF1 24940 95 - 1 CAUCG--------------GCGUUUUU-A-UUUUAUUUUCAUUUGCG--------CGCUGCCGGGAGAGACAAAAUUUAAAUUGUUUUUCCCAUUGGCAUCGGCA-GAAAACAAAUUCUA .....--------------........-.-...........(((((.--------((.(((((((((((((((........)))))))))))...)))).)))))-))............ ( -27.10) >DroYak_CAF1 25090 95 - 1 CAUCG--------------ACGUUUUU-A-UUUUAUUUUCAUUUGCG--------CUCUGCCAGCAGAGACAAAAUUUAAAUUGUUUUUCCCAUUGGCAUCGGCG-GAAAACAAAUUCUA .....--------------........-.-...........(((.((--------((.((((((.((((((((........))))))))....))))))..))))-.))).......... ( -18.40) >DroAna_CAF1 19077 102 - 1 CAUCCCCGCCAUCGAGGCAACAUUUUU-A-UUUUAUUUUCAUUUUC---------UACUGCCAGCAGAGACAACAUUUAAAUUGUUUUUGCC------AUCGGCG-GAAAACAAAUUCUA .....(((((.....((((........-.-................---------...)))).((((((((((........)))))))))).------...))))-)............. ( -24.79) >DroPer_CAF1 71292 99 - 1 CAUCG--------------ACGAUUUUCAUUUUUAUUUUCAUUUUCUGUUGCUGUUGCUGUUGGCAGAGACAAAAUUUAAAUUGUUUUUGCC------AACGGCU-GAAAGCAAAUUCUA .....--------------.............................(((((...(((((((((((((((((........)))))))))))------)))))).-...)))))...... ( -32.00) >consensus CAUCG______________ACGUUUUU_A_UUUUAUUUUCAUUUGCG________CGCUGCCAGCAGAGACAAAAUUUAAAUUGUUUUUCCC______AUCGGCG_GAAAACAAAUUCUA ...........................................................(((.((((((((((........))))))))))..........)))................ ( -9.01 = -8.82 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:48 2006