
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,506,532 – 20,506,661 |
| Length | 129 |
| Max. P | 0.798024 |

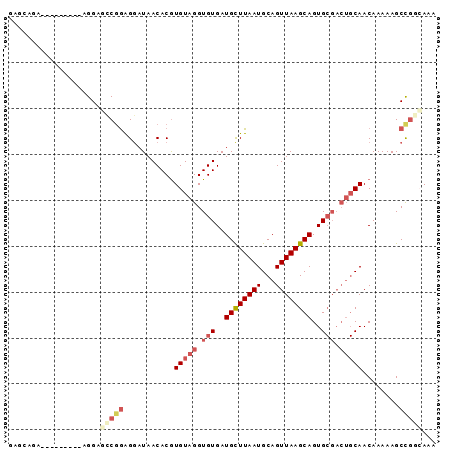
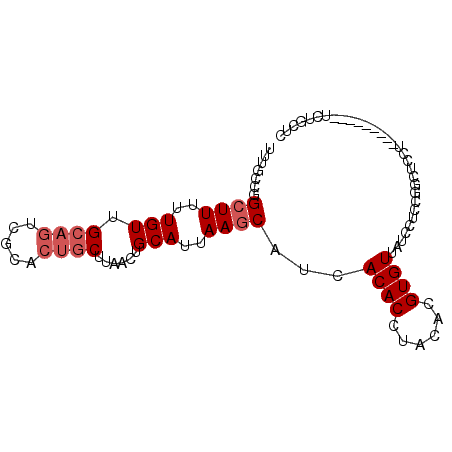
| Location | 20,506,532 – 20,506,625 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 80.55 |
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -14.19 |
| Energy contribution | -16.07 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20506532 93 + 22407834 GAGCAGAAGGGGCCGCAGGAGCCGGAGGAUAACACGUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGACUGCAACAAAAAGCCGGCAAA ..((.(......).))....(((((...........(((((.(((..((((((((...))))))))..))).)))))........)))))... ( -26.51) >DroSec_CAF1 23571 84 + 1 GAGCAGA---------AGGAGUCGGAGGAUAACACGUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGACUGCAACAAAAAGCCGGAAAA .......---------.....((((...........(((((.(((..((((((((...))))))))..))).)))))........)))).... ( -19.11) >DroSim_CAF1 28670 84 + 1 GAGCAGA---------AGGAGCCGGAGGAUAACACGUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGACUGCAACAAAAAGCCGGCAAA .......---------....(((((...........(((((.(((..((((((((...))))))))..))).)))))........)))))... ( -25.71) >DroYak_CAF1 22426 77 + 1 ----------------UGGAUCCGGAGGAUAACACAUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGACUGCAACAAAAAGCUGCAAAA ----------------........(((.((.((((......)))).)).)))..(((((((..((((.....)))).......)))))))... ( -18.60) >DroAna_CAF1 14694 71 + 1 ----------------AGGCUGCAAGGACUAACACGUGUAGGUGUGAUGUUUAAUGCAGUUAAGCAGUG------CAACAAAAAGCCAGCAAA ----------------.(((((((.((((..((((......))))...))))..)))))))..((.(.(------(........))).))... ( -17.80) >consensus GAGCAGA_________AGGAGCCGGAGGAUAACACGUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGACUGCAACAAAAAGCCGGCAAA ....................(((((...........(((((.(((..((((((((...))))))))..))).)))))........)))))... (-14.19 = -16.07 + 1.88)


| Location | 20,506,532 – 20,506,625 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 80.55 |
| Mean single sequence MFE | -17.64 |
| Consensus MFE | -9.92 |
| Energy contribution | -10.72 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20506532 93 - 22407834 UUUGCCGGCUUUUUGUUGCAGUCGCACUGCUUAACUGCAUUAAGCAUCACACCUACACGUGUUAUCCUCCGGCUCCUGCGGCCCCUUCUGCUC ...((((((((..(((.((((.....))))......)))..))))...((((......)))).......))))....((((......)))).. ( -24.20) >DroSec_CAF1 23571 84 - 1 UUUUCCGGCUUUUUGUUGCAGUCGCACUGCUUAACUGCAUUAAGCAUCACACCUACACGUGUUAUCCUCCGACUCCU---------UCUGCUC .....((((((..(((.((((.....))))......)))..))))...((((......)))).......))......---------....... ( -14.50) >DroSim_CAF1 28670 84 - 1 UUUGCCGGCUUUUUGUUGCAGUCGCACUGCUUAACUGCAUUAAGCAUCACACCUACACGUGUUAUCCUCCGGCUCCU---------UCUGCUC ...((((((((..(((.((((.....))))......)))..))))...((((......)))).......))))....---------....... ( -21.00) >DroYak_CAF1 22426 77 - 1 UUUUGCAGCUUUUUGUUGCAGUCGCACUGCUUAACUGCAUUAAGCAUCACACCUACAUGUGUUAUCCUCCGGAUCCA---------------- ..(((((((.....)))))))......(((((((.....))))))).((((......))))..((((...))))...---------------- ( -16.10) >DroAna_CAF1 14694 71 - 1 UUUGCUGGCUUUUUGUUG------CACUGCUUAACUGCAUUAAACAUCACACCUACACGUGUUAGUCCUUGCAGCCU---------------- ...((..((........)------)...))....(((((........(((........)))........)))))...---------------- ( -12.39) >consensus UUUGCCGGCUUUUUGUUGCAGUCGCACUGCUUAACUGCAUUAAGCAUCACACCUACACGUGUUAUCCUCCGGCUCCU_________UCUGCUC .......((((..(((.((((.....))))......)))..))))...((((......))))............................... ( -9.92 = -10.72 + 0.80)

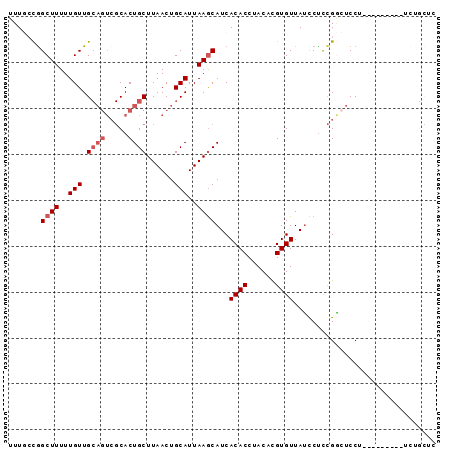
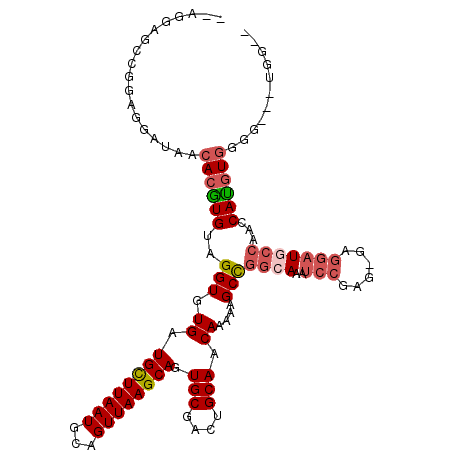
| Location | 20,506,548 – 20,506,661 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -30.85 |
| Consensus MFE | -20.34 |
| Energy contribution | -21.33 |
| Covariance contribution | 0.99 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20506548 113 + 22407834 --AGGAGCCGGAGGAUAACACGUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGACUGCAACAAAAAGCCGGCAAAUCCGAGAGAGGAUGCCAACCAUGUGGGGCGGUGGGU --....(((.............(((((.(((..((((((((...))))))))..))).))))).......(((((((..(((......)))))))..((....)))))))).... ( -35.30) >DroSec_CAF1 23578 113 + 1 --AGGAGUCGGAGGAUAACACGUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGACUGCAACAAAAAGCCGGAAAAUCCGAGUGAGGAUGCCAACCAUGUGGGACGGUGGUA --.(.(((((.(.....((((......))))..((((((((...)))))))).).))))).)........(((((...((((......)))).))..((....))...))).... ( -31.30) >DroSim_CAF1 28677 113 + 1 --AGGAGCCGGAGGAUAACACGUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGACUGCAACAAAAAGCCGGCAAAUCCGAGUGAGGAUGCCAACCAUGUGGGACGGUGGUA --.((.(((((...........(((((.(((..((((((((...))))))))..))).)))))........)))))..((((......)))).)).(((((........))))). ( -35.91) >DroEre_CAF1 22407 104 + 1 UGAGGAUCCGGAGGAUAACACGUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGAAUGCAACAAAAAGCCGGCAAAUCC-A--GAGGAUGCCACCCAUGUGGGG-------- ...((((((...((((......(((.(((.((.((((((((...)))))))).(((....))).))....))).))).))))-.--..)))).)).(((....))).-------- ( -30.00) >DroYak_CAF1 22426 103 + 1 --UGGAUCCGGAGGAUAACACAUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGACUGCAACAAAAAGCUGCAAAAUCCGA--GAGGAUGCCAACCAUGUGGGG-------- --(((((((...((((..(((((....)))))........(((((((..((((.....)))).......)))))))..))))..--..)))).))).((....))..-------- ( -30.40) >DroAna_CAF1 14694 103 + 1 --AGGCUGCAAGGACUAACACGUGUAGGUGUGAUGUUUAAUGCAGUUAAGCAGUG------CAACAAAAAGCCAGCAAAUCCAAAGGAUUCUGCCACACACAUAUA----UAAAU --.(((((((.((((..((((......))))...))))..)))))))..((((((------(............)))(((((...)))))))))............----..... ( -22.20) >consensus __AGGAGCCGGAGGAUAACACGUGUAGGUGUGAUGCUUAAUGCAGUUAAGCAGUGCGACUGCAACAAAAAGCCGGCAAAUCCGAG_GAGGAUGCCAACCAUGUGGGG___UGG__ ..................((((((..(((.((.((((((((...)))))))).(((....))).))....)))((((..(((......)))))))...))))))........... (-20.34 = -21.33 + 0.99)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:45 2006