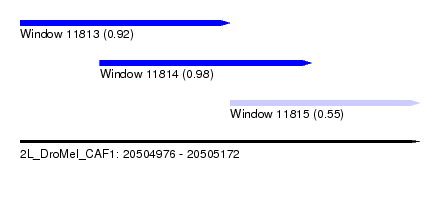
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,504,976 – 20,505,172 |
| Length | 196 |
| Max. P | 0.977028 |

| Location | 20,504,976 – 20,505,079 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -25.97 |
| Energy contribution | -27.25 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
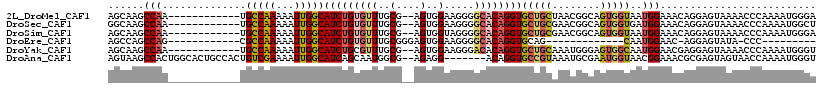
>2L_DroMel_CAF1 20504976 103 + 22407834 UACCGCUAACUGCGCAUACAAUGGCUGCGGAA-AUUCCCCAGCAAGCCAA------------UGCCAAAAAUUGGCAUCUGUGUUUGCG--AGUGGAAGGGGCACAGGUGCUGCUAAC ..(((((...((((.(((((.(((((((((..-.....)).)).)))))(------------(((((.....)))))).))))).))))--))))).((.((((....)))).))... ( -38.10) >DroSec_CAF1 22061 103 + 1 UACCGCUAACUGCGCAUACAAUGGCUGUGGAA-AUUCCCCGGCAAGCCAA------------UGCCAAAAAUUGGCAUCUGUGUUUGCG--AGUGGAAGGGGCACAGGUGCUGCGAAC ..(((((...((((.(((((...((((.((..-...)).))))..(((((------------(.......))))))...))))).))))--)))))..(.((((....)))).).... ( -36.20) >DroSim_CAF1 27128 103 + 1 UACCGCUAACUGCGCAUACAAUGGCUGUGGAA-AUUCCCCAGCAAGCCAA------------UGCCAAAAAUUGGCAUCUGUGUUUGCG--AGUGGUAGGGGCACAGGUGCUGCGAAC (((((((...((((.(((((...((((.((..-...)).))))..(((((------------(.......))))))...))))).))))--)))))))(.((((....)))).).... ( -39.10) >DroEre_CAF1 20945 101 + 1 UACCGCUAACUGCGCAUACAAUGGCUGUGGAAAAUUCCCCAGCCAGCCAG------------CGCCAAAAAUUGGCAUCUGUGUUUGCGGGAGUGGAAGGGGCACAGGUGCAG----- ..(((((..(((((.(((((.((((((.(((....))).)))))).....------------.((((.....))))...))))).))))).))))).....(((....)))..----- ( -39.60) >DroYak_CAF1 20880 104 + 1 UACCGCUAACUGCGCUUACAAUGGCUGUGGAAAAUGGCCCAGCAAGCCAA------------UGCCAAAAAUUGGCAUCUGCGUUUGCG--AGUGGAAGGGACACAGGUGCUGCAAAU ....((.....((((((.....((((((.....))))))..(((((((((------------(((((.....)))))).)).)))))).--.(((.......))))))))).)).... ( -34.30) >DroAna_CAF1 13453 109 + 1 UACCGCUAACUGCAAUAGCAAUAGCUGCGAAAAAUUCGGUAGUAAGCCACUGGCACUGCCACUGUCGAAAAUUGGCAUCAGCAAUGGCG--AGAGG-------ACAGGUGCCGUAAAU (((.(((.(((((..((((....))))(((.....)))))))).)))....((((((....((.(((...((((.......))))..))--).)).-------...)))))))))... ( -33.90) >consensus UACCGCUAACUGCGCAUACAAUGGCUGUGGAA_AUUCCCCAGCAAGCCAA____________UGCCAAAAAUUGGCAUCUGUGUUUGCG__AGUGGAAGGGGCACAGGUGCUGCAAAC ..(((((...((((.(((((.(((((((((........)).)).)))))..............(((((...)))))...))))).))))..)))))..(.((((....)))).).... (-25.97 = -27.25 + 1.28)
| Location | 20,505,015 – 20,505,119 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.54 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -15.50 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
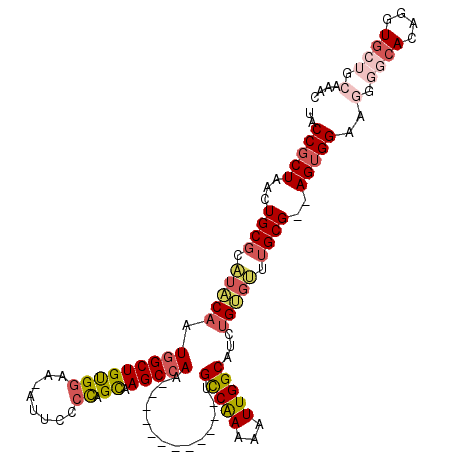
>2L_DroMel_CAF1 20505015 104 + 22407834 AGCAAGCCAA------------UGCCAAAAAUUGGCAUCUGUGUUUGCG--AGUGGAAGGGGCACAGGUGCUGCUAACGGCAGUGGUAAUGGAAACAGGAGUAAAACCCAAAAUGGGA .((..((((.------------((((.......(((((((((((((.(.--.......))))))))))))))......)))).))))..((....))...))....(((.....))). ( -35.52) >DroSec_CAF1 22100 104 + 1 GGCAAGCCAA------------UGCCAAAAAUUGGCAUCUGUGUUUGCG--AGUGGAAGGGGCACAGGUGCUGCGAACGGCAGUGGUGAUGGAAACAGGAGUAAAACCCAAAAUGGCU .(((((((((------------(((((.....)))))).)).)))))).--.......(((.(((.(.(((((....))))).).))).((....)).........)))......... ( -34.20) >DroSim_CAF1 27167 104 + 1 AGCAAGCCAA------------UGCCAAAAAUUGGCAUCUGUGUUUGCG--AGUGGUAGGGGCACAGGUGCUGCGAACGGCAGUGGUAAUGGAAACAGGAGUAAAACCCAAAAUGGGA .((..((((.------------((((.......(((((((((((((((.--....))))..)))))))))))......)))).))))..((....))...))....(((.....))). ( -36.52) >DroEre_CAF1 20985 82 + 1 AGCCAGCCAG------------CGCCAAAAAUUGGCAUCUGUGUUUGCGGGAGUGGAAGGGGCACAGGUGCAG-------------CAAUGGAAC-AGGAGUAUA-CCC--------- ..((..(((.------------.((((.....))(((((((((((..(....)..).....)))))))))).)-------------)..)))...-.))......-...--------- ( -23.30) >DroYak_CAF1 20920 104 + 1 AGCAAGCCAA------------UGCCAAAAAUUGGCAUCUGCGUUUGCG--AGUGGAAGGGACACAGGUGCUGCAAAUGGGAGUGGCAAUGGAACGAGGAGUAAAACCCAAAAUGGGU .(((((((((------------(((((.....)))))).)).)))))).--.(((.......)))...(((..(........)..))).................((((.....)))) ( -30.60) >DroAna_CAF1 13493 109 + 1 AGUAAGCCACUGGCACUGCCACUGUCGAAAAUUGGCAUCAGCAAUGGCG--AGAGG-------ACAGGUGCCGUAAAUGCGAAUGGUAACGGAAACGCGAGUAGUAACCAAAAUGGGU .....(((...((((((....((.(((...((((.......))))..))--).)).-------...))))))...........((((.((....((....)).)).)))).....))) ( -29.90) >consensus AGCAAGCCAA____________UGCCAAAAAUUGGCAUCUGUGUUUGCG__AGUGGAAGGGGCACAGGUGCUGCAAACGGCAGUGGUAAUGGAAACAGGAGUAAAACCCAAAAUGGGU ......(((..............(((((...)))))(((((((((..(....)..).....))))))))(((((........)))))..))).......................... (-15.50 = -16.10 + 0.60)
| Location | 20,505,079 – 20,505,172 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 85.62 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -20.47 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
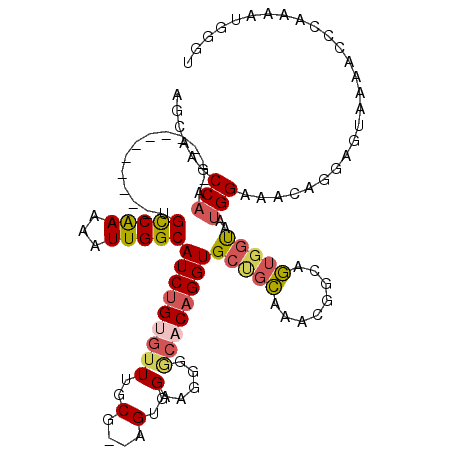
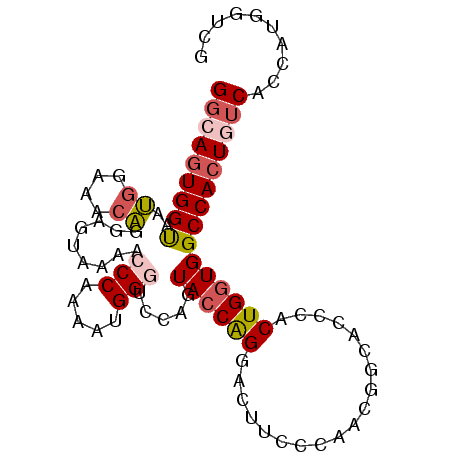
>2L_DroMel_CAF1 20505079 93 + 22407834 GGCAGUGGUAAUGGAAACAGGAGUAAAACCCAAAAUGGGACCAGUACCAGGACAUC-CAACGGCACCCACUGGUGGCCACUGUCACCAUGGUCG (((((((((..((....)).........(((.....)))((((((....((....)-)...((...)))))))).))))))))).......... ( -32.20) >DroSec_CAF1 22164 94 + 1 GGCAGUGGUGAUGGAAACAGGAGUAAAACCCAAAAUGGCUCCAGUACCAGGACUUCCCAACGGCACCCACUGGUGGCCACUGUCACCAUGGUCG (((.((((((((((.....((........))....((((((((((....((.....))...((...))))))).))))))))))))))).))). ( -34.60) >DroSim_CAF1 27231 94 + 1 GGCAGUGGUAAUGGAAACAGGAGUAAAACCCAAAAUGGGACCAGUACCAGGACUUCCCAACGGCACCCACUGGUGGCCACUGUCACCAUGGUCG (((((((((..((....)).........(((.....)))((((((....((.....))...((...)))))))).))))))))).......... ( -31.60) >DroYak_CAF1 20984 94 + 1 GGGAGUGGCAAUGGAACGAGGAGUAAAACCCAAAAUGGGUCCAGUACCAGGACUUUCAAACAGCAUCCACUGGUGGCCACUGUCACCAUGGUCG ((.((((((..((((((.....))....(((.....))))))).((((((...................)))))))))))).)).......... ( -27.71) >DroAna_CAF1 13562 93 + 1 GCGAAUGGUAACGGAAACGCGAGUAGUAACCAAAAUGGGUGCAGUAACGGGAGUUG-GAAUGGCAGCCACUGGUGGCCACUGUCACCACGAAAG .((..((((.((....((....)).)).)))).....(((((((((((....))))-...((((.(((...))).))))))).)))).)).... ( -28.20) >consensus GGCAGUGGUAAUGGAAACAGGAGUAAAACCCAAAAUGGGUCCAGUACCAGGACUUCCCAACGGCACCCACUGGUGGCCACUGUCACCAUGGUCG (((((((((..((....)).........(((.....))).....((((((...................))))))))))))))).......... (-20.47 = -21.43 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:43 2006