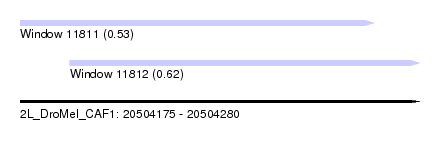
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,504,175 – 20,504,280 |
| Length | 105 |
| Max. P | 0.616744 |

| Location | 20,504,175 – 20,504,268 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.03 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -21.35 |
| Energy contribution | -21.02 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
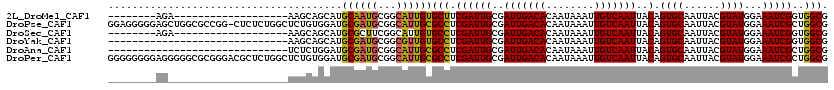
>2L_DroMel_CAF1 20504175 93 + 22407834 --------AGA-------------------AAGCAGCAUGCAAUGCGGCAUUGUGCUUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGGUGGCG --------...-------------------..((.((((((((((.(((.....))).).)))))((((((((........))))))))...)))).....((.((....))))...)). ( -23.60) >DroPse_CAF1 60383 119 + 1 GGAGGGGGAGCUGGCGCCGG-CUCUCUGGCUCUGUGGAUGCGAUGCGGCAUUGCGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGCUGGCG ....(((((((((....)))-)))))).(((..((((...(.(((((..(((((((......((..(((((((........)))))))..)))))))))..))))).)...)))).))). ( -45.70) >DroSec_CAF1 21292 93 + 1 --------AGA-------------------AAGCAGCAUGCGCUUCGGCAUUGUGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGGUGGCG --------...-------------------..((.....))((....)).....((((((((((..(((((((........)))))))..).((((......))))...)))))).))). ( -26.50) >DroYak_CAF1 20139 90 + 1 ------------------------------AAGCAGCAUGCGAUGCGGCGUUGUGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGGUGGCG ------------------------------..(((((.(((...)))..)))))((((((((((..(((((((........)))))))..).((((......))))...)))))).))). ( -26.30) >DroAna_CAF1 12648 90 + 1 ------------------------------UCUCUGGAUGCGAUGCGGCAUUGCGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGCUGGCG ------------------------------...(..(...(.(((((..(((((((......((..(((((((........)))))))..)))))))))..))))).)......)..).. ( -24.80) >DroPer_CAF1 66162 120 + 1 GGGGGGGGAGGGGGCGCGGGACGCUCUGGCUCUGUGGAUGCGAUGCGGCAUUGCGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGCUGGCG ....((((..((((((.....))))))..))))((((...(.(((((..(((((((......((..(((((((........)))))))..)))))))))..))))).)...))))..... ( -40.00) >consensus ________AG____________________AAGCAGCAUGCGAUGCGGCAUUGCGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGCUGGCG .......................................((((((...))))))(((.((((((..(((((((........)))))))..).((((......))))...)))))..))). (-21.35 = -21.02 + -0.33)


| Location | 20,504,188 – 20,504,280 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -20.05 |
| Energy contribution | -20.13 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20504188 92 + 22407834 CAAUGCGGCAUUGUGCUUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGGUGGCGUUCUCU----------------------------UUCCAC ..((((((((((((((.......))((((((((........))))))))))))))).....)))))(((((..((......))..)----------------------------)))).. ( -24.10) >DroPse_CAF1 60422 120 + 1 CGAUGCGGCAUUGCGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGCUGGCGUUCCCGCAGGGUCUGGUGCGGGGGGCCUACCCCCUGGCAG ...(((((....(((((.((((((..(((((((........)))))))..).((((......))))...)))))..)))))..)))))........(((((((((....)))))).))). ( -45.50) >DroSec_CAF1 21305 92 + 1 CGCUUCGGCAUUGUGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGGUGGCGUUCUCU----------------------------GUCCAC .((....)).....((((((((((..(((((((........)))))))..).((((......))))...)))))).))).......----------------------------...... ( -25.00) >DroEre_CAF1 20215 91 + 1 CGAUGCGGCAUUGUGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGGUGGCGU-CUCU----------------------------GUGCAC ...((((.((..((((((((((((..(((((((........)))))))..).((((......))))...)))))).)))).-)..)----------------------------))))). ( -25.90) >DroAna_CAF1 12658 86 + 1 CGAUGCGGCAUUGCGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGCUGGCGUUCAC----------------------------------C ......((....(((((.((((((..(((((((........)))))))..).((((......))))...)))))..)))))...)----------------------------------) ( -23.20) >DroPer_CAF1 66202 108 + 1 CGAUGCGGCAUUGCGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGCUGGCGUUCCCGCAGGGUCUGGUGCGGGGGGC-UA----------- (.(((((..(((((((......((..(((((((........)))))))..)))))))))..))))).).......((((.((((((((........))))))))))-))----------- ( -38.40) >consensus CGAUGCGGCAUUGCGCCUCGAUUGCGAUUGACACAAUAAAUUGUCAAUUACAGUGCAAUUACGUAUGGAAAUCGCUGGCGUUCUCU_____________________________U_CAC (.(((((..(((((((......((..(((((((........)))))))..)))))))))..))))).).................................................... (-20.05 = -20.13 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:40 2006