
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,499,791 – 20,499,906 |
| Length | 115 |
| Max. P | 0.865096 |

| Location | 20,499,791 – 20,499,906 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.86 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
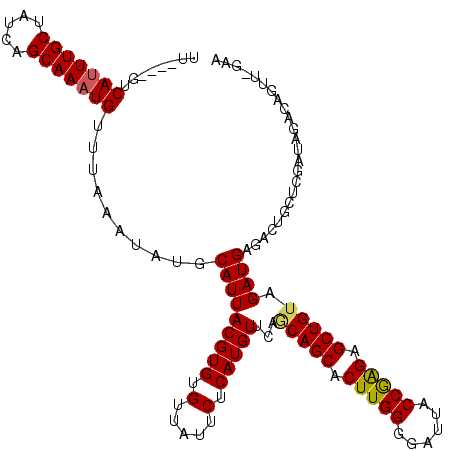
>2L_DroMel_CAF1 20499791 115 + 22407834 UUC-AACUGUCUAUCGAGCAGUCUCAUCUACAGCUCUCGGUAAUCCCCAAGUGCUGCUGAACAUGAGAAUAACACACGUAAUGCAUAUUUAAACAUUUGCUGAUAGCAAAUGAC----AA .((-(..((.(((((.((((((((((((..((((.((.((......)).)).))))..))...)))))...((....)).................))))))))))))..))).----.. ( -26.70) >DroPse_CAF1 56112 100 + 1 --------------------CUUUCAUCUACAGCUCUCGGUAAUCCCCAAGUGCUGCUCAACAUGGGAAUAACACACGUAAUGCAUAUUUAAACAUUUGCUGAUGGCAAAUGACACACAA --------------------.(..(((...((((.((.((......)).)).))))......)))..).........................(((((((.....)))))))........ ( -18.60) >DroSec_CAF1 16766 115 + 1 UUC-AACUGUAUAUCGAGCAGUCUCAUCUACAGCUCUCGGUAAUCCCCAAGUGCUGUUGAACAUGAGAAUAACACACGUAAUGCAUAUUUAAACAUUUGCUGAUAGCAAAUGAC----AA ...-...(((((..((.....(((((((.(((((.((.((......)).)).))))).))...)))))........))..)))))........(((((((.....)))))))..----.. ( -27.62) >DroYak_CAF1 15961 115 + 1 UUU-CAAUGUCUAUCGAGCAGUCACAUCUACAGCUCUUGGUAAUCCCCAAGUGCUGCUGAACAUGAGAAUAACACACGUAAUGCAUAUUUAAACAUUUGCUGAUAGCAAGUGAC----AA ..(-((.((.(((((.(((((.....((..((((.(((((......))))).))))..))..(((...((.((....)).)).)))..........))))))))))))..))).----.. ( -27.00) >DroAna_CAF1 8602 116 + 1 UUCCAACUCUCUAUCGAGGACUCUCAUCGACAGCUCCUGGUAAUCCCCAAGUGCUGAUGAACAUGAGAAUAACACACGUAAUGCAUAUUUAAACAUUUGCUGAUAGCAAAUGAC----AA ......(((......)))...((((((((.((((.(.(((......))).).)))).)))...))))).........................(((((((.....)))))))..----.. ( -26.20) >DroPer_CAF1 62020 100 + 1 --------------------CUUUCAUCUACAGCUCUCGGUAAUCCCCAAGUGCUGCUGAACAUGGGAAUAACACACGUAAUGCAUAUUUAAACAUUUGCUGAUGGCAAAUGACACACAA --------------------.(..((((..((((.((.((......)).)).))))..))...))..).........................(((((((.....)))))))........ ( -20.00) >consensus UUC_AACUGUCUAUCGAGCACUCUCAUCUACAGCUCUCGGUAAUCCCCAAGUGCUGCUGAACAUGAGAAUAACACACGUAAUGCAUAUUUAAACAUUUGCUGAUAGCAAAUGAC____AA .....................((((((...((((.((.((......)).)).))))......)))))).........................(((((((.....)))))))........ (-19.88 = -19.68 + -0.19)



| Location | 20,499,791 – 20,499,906 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.86 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -26.04 |
| Energy contribution | -25.90 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20499791 115 - 22407834 UU----GUCAUUUGCUAUCAGCAAAUGUUUAAAUAUGCAUUACGUGUGUUAUUCUCAUGUUCAGCAGCACUUGGGGAUUACCGAGAGCUGUAGAUGAGACUGCUCGAUAGACAGUU-GAA ..----.(((.((((((((((((........(((((((.....)))))))..((((((.(...(((((.(((((......))))).)))))).)))))).)))).))))).))).)-)). ( -38.80) >DroPse_CAF1 56112 100 - 1 UUGUGUGUCAUUUGCCAUCAGCAAAUGUUUAAAUAUGCAUUACGUGUGUUAUUCCCAUGUUGAGCAGCACUUGGGGAUUACCGAGAGCUGUAGAUGAAAG-------------------- .((((((((((((((.....))))))).....)))))))..(((((.(.....).)))))...(((((.(((((......))))).))))).........-------------------- ( -27.40) >DroSec_CAF1 16766 115 - 1 UU----GUCAUUUGCUAUCAGCAAAUGUUUAAAUAUGCAUUACGUGUGUUAUUCUCAUGUUCAACAGCACUUGGGGAUUACCGAGAGCUGUAGAUGAGACUGCUCGAUAUACAGUU-GAA ..----.(((.(((.((((((((........(((((((.....)))))))..((((((.(...(((((.(((((......))))).)))))).)))))).)))).))))..))).)-)). ( -33.50) >DroYak_CAF1 15961 115 - 1 UU----GUCACUUGCUAUCAGCAAAUGUUUAAAUAUGCAUUACGUGUGUUAUUCUCAUGUUCAGCAGCACUUGGGGAUUACCAAGAGCUGUAGAUGUGACUGCUCGAUAGACAUUG-AAA ..----.(((..(((((((((((........(((((((.....)))))))....(((((((..(((((.(((((......))))).))))).))))))).)))).))))).)).))-).. ( -35.70) >DroAna_CAF1 8602 116 - 1 UU----GUCAUUUGCUAUCAGCAAAUGUUUAAAUAUGCAUUACGUGUGUUAUUCUCAUGUUCAUCAGCACUUGGGGAUUACCAGGAGCUGUCGAUGAGAGUCCUCGAUAGAGAGUUGGAA ..----..(((((((.....)))))))....(((((((.....)))))))(((((((...((..((((.(((((......))))).))))..))))))))).(((....)))........ ( -34.40) >DroPer_CAF1 62020 100 - 1 UUGUGUGUCAUUUGCCAUCAGCAAAUGUUUAAAUAUGCAUUACGUGUGUUAUUCCCAUGUUCAGCAGCACUUGGGGAUUACCGAGAGCUGUAGAUGAAAG-------------------- .((((((((((((((.....))))))).....)))))))..(((((.(.....).)))))((.(((((.(((((......))))).))))).))......-------------------- ( -28.50) >consensus UU____GUCAUUUGCUAUCAGCAAAUGUUUAAAUAUGCAUUACGUGUGUUAUUCUCAUGUUCAGCAGCACUUGGGGAUUACCGAGAGCUGUAGAUGAGACUGCUCGAUAGACAGUU_GAA ........(((((((.....)))))))..........(((((((((.(.....).)))))...(((((.(((((......))))).))))).))))........................ (-26.04 = -25.90 + -0.14)

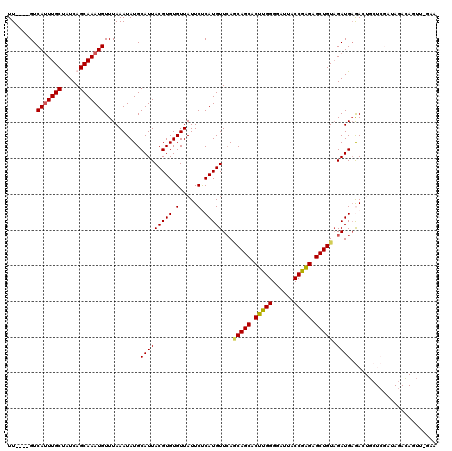
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:35 2006