
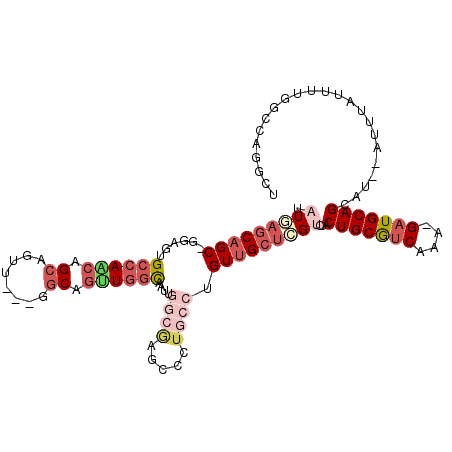
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,492,931 – 20,493,037 |
| Length | 106 |
| Max. P | 0.913447 |

| Location | 20,492,931 – 20,493,037 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -23.02 |
| Energy contribution | -25.38 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
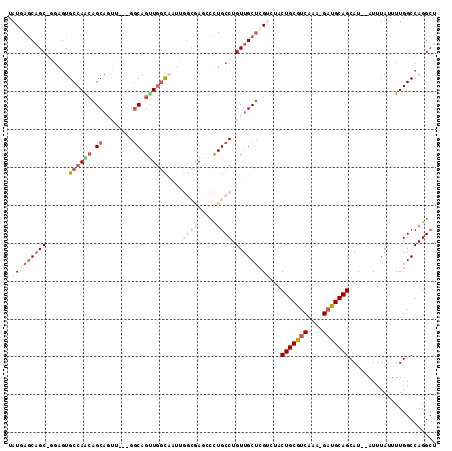
>2L_DroMel_CAF1 20492931 106 + 22407834 UAUGAGCAGC-GGAGUGCCAACAGCAGUU---GGCAGUUGGCAAUUGGCAAGCCCUGCCUGUUGCUCGUCUGCUGCGUCAAA-GAUGCAGCAU--AUUUAUUUUGGCCAGGCU .(((((((((-(((.(((((((....)))---)))).))((((...((....)).)))))))))))))).(((((((((...-))))))))).--.................. ( -44.60) >DroSec_CAF1 9993 106 + 1 UAUUAGCAGC-GGAGUGCCAACAGCAGUU---GGCAGCUGGCAAUUGGCGAGCCCUGCCUGUUGCUCGUCUACUGCGUCAGA-GAUGCAGCAU--AUUUAUUUUGGCCAGGCU .......(((-....(((((((....)))---)))).(((((....(((((((..........)))))))..(((((((...-)))))))...--..........)))))))) ( -40.10) >DroSim_CAF1 15116 106 + 1 UAUGAGCAGC-GGAGUGCCAACAGCAGUU---GGCAGUUGGCAAUUGGCGAGUCCUGCCUGUUGCUCGUCUACUGCGUCAGA-GAUGCAGCAU--AUUUAUUUUGGCCAGGCU ...(((((((-.((.(((((((....)))---)))).)).))....((((.....))))...)))))((((.(((((((...-)))))))((.--........))...)))). ( -39.50) >DroYak_CAF1 9159 106 + 1 UAUGAGCAGC-GAAGUGCCAACGGCAUAU---GGCAGUUGGCAAUUGGCAAACCCUGCCUGUUGCUCGUCUUCUGCGUCAAA-GAUGCAGCAU--AUUUAUUUUGGCCAGGCU ...(((((((-(...(((((((.((....---.)).)))))))...((((.....))))))))))))((((.(((((((...-)))))))((.--........))...)))). ( -39.00) >DroAna_CAF1 1950 95 + 1 UAUAAGCAGC-CGAGUGCCAGGAGCAGUU---GGCAGUUUGU-------GA-ACUUGCCUGUUGCUUG---ACUGCAUCAAA-GAUGCAGCAU--AUUUAUUUUGGCCAGGAA (((((((.((-(((.(((.....))).))---))).))))))-------).-.((.(((.((....((---.(((((((...-))))))))).--....))...))).))... ( -32.50) >DroPer_CAF1 54081 102 + 1 UAUAAUCUGCUCGACUGCCAGCUGCCUCGACUGCCAGCUGCCU--------CUGCUGCCUGUUGCACG---ACUGCAUCAAACGUUGCAGCAUAUAUUUAUUUUGGCCAGGCA ........((..(...(((((((((..((...(.((((.....--------..)))).)((.((((..---..)))).))..))..)))))(((....)))..)))))..)). ( -25.80) >consensus UAUGAGCAGC_GGAGUGCCAACAGCAGUU___GGCAGUUGGCAAUUGGCGAGCCCUGCCUGUUGCUCGUCUACUGCGUCAAA_GAUGCAGCAU__AUUUAUUUUGGCCAGGCU .(((((((((......((((((.((........)).))))))....((((.....)))).)))))))))...(((((((....)))))))....................... (-23.02 = -25.38 + 2.36)

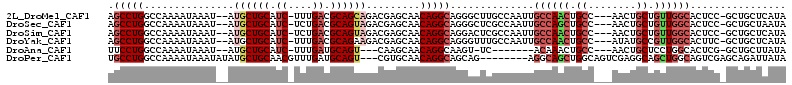
| Location | 20,492,931 – 20,493,037 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -18.92 |
| Energy contribution | -20.37 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
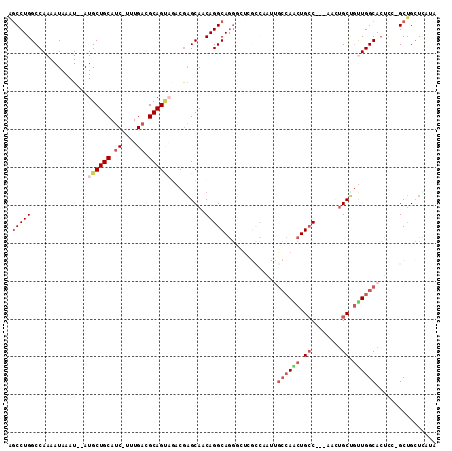
>2L_DroMel_CAF1 20492931 106 - 22407834 AGCCUGGCCAAAAUAAAU--AUGCUGCAUC-UUUGACGCAGCAGACGAGCAACAGGCAGGGCUUGCCAAUUGCCAACUGCC---AACUGCUGUUGGCACUCC-GCUGCUCAUA .(((((............--.((((((.((-...)).)))))).........))))).((((..((....(((((((.((.---....)).)))))))....-)).))))... ( -37.95) >DroSec_CAF1 9993 106 - 1 AGCCUGGCCAAAAUAAAU--AUGCUGCAUC-UCUGACGCAGUAGACGAGCAACAGGCAGGGCUCGCCAAUUGCCAGCUGCC---AACUGCUGUUGGCACUCC-GCUGCUAAUA (((((.(((.........--.((((((.((-...)).))))))...........))).))))).....((((.((((((((---(((....)))))))....-)))).)))). ( -35.70) >DroSim_CAF1 15116 106 - 1 AGCCUGGCCAAAAUAAAU--AUGCUGCAUC-UCUGACGCAGUAGACGAGCAACAGGCAGGACUCGCCAAUUGCCAACUGCC---AACUGCUGUUGGCACUCC-GCUGCUCAUA ..................--.((((((.((-...)).))))))...(((((.(.(((.......)))...(((((((.((.---....)).)))))))....-).)))))... ( -32.20) >DroYak_CAF1 9159 106 - 1 AGCCUGGCCAAAAUAAAU--AUGCUGCAUC-UUUGACGCAGAAGACGAGCAACAGGCAGGGUUUGCCAAUUGCCAACUGCC---AUAUGCCGUUGGCACUUC-GCUGCUCAUA .(((((............--.((((.(.((-(((......))))).))))).))))).((((..((.((.(((((((.((.---....)).))))))).)).-)).))))... ( -34.24) >DroAna_CAF1 1950 95 - 1 UUCCUGGCCAAAAUAAAU--AUGCUGCAUC-UUUGAUGCAGU---CAAGCAACAGGCAAGU-UC-------ACAAACUGCC---AACUGCUCCUGGCACUCG-GCUGCUUAUA .....((((.........--....((((((-...))))))((---((((((...((((.((-(.-------...)))))))---...))))..))))....)-)))....... ( -26.30) >DroPer_CAF1 54081 102 - 1 UGCCUGGCCAAAAUAAAUAUAUGCUGCAACGUUUGAUGCAGU---CGUGCAACAGGCAGCAG--------AGGCAGCUGGCAGUCGAGGCAGCUGGCAGUCGAGCAGAUUAUA (((.((((.............((((((.....(((.((((..---..)))).))))))))).--------.(.((((((.(......).)))))).).)))).)))....... ( -33.50) >consensus AGCCUGGCCAAAAUAAAU__AUGCUGCAUC_UUUGACGCAGUAGACGAGCAACAGGCAGGGCUCGCCAAUUGCCAACUGCC___AACUGCUGUUGGCACUCC_GCUGCUCAUA .(((((...............((((((.((....)).)))))).........)))))..............((((((.((........)).))))))................ (-18.92 = -20.37 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:31 2006