
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,490,346 – 20,490,553 |
| Length | 207 |
| Max. P | 0.991391 |

| Location | 20,490,346 – 20,490,455 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.27 |
| Mean single sequence MFE | -46.14 |
| Consensus MFE | -37.84 |
| Energy contribution | -37.80 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20490346 109 + 22407834 AUCUGACUGCAUUUUCCAAUUGCUCCGGAAUUUGGUGGCCA-----AGUCCGGGAGGCGUCCCUUGUGGUGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGC .((((...(((.........)))..))))((((((...)))-----)))((((.(((((((((....)).)))))))(((((...(((((((....))))))).))))))))). ( -45.70) >DroSec_CAF1 7394 109 + 1 AUCUGACUGCAUUUUCCAAUCGCCCUGGAAUUCGGUGGCCA-----AGUCCGGGAGGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGC ...((.(..(...(((((.......)))))....)..).))-----...((((.(((((((((....)).)))))))(((((...(((((((....))))))).))))))))). ( -46.30) >DroSim_CAF1 12537 108 + 1 AUCUGACUGCAUUUUCCAAUCGCCCCGGAAUUCG-UGGGCA-----AGUCCGGGAGGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGC .....................((((((.....))-.)))).-----...((((.(((((((((....)).)))))))(((((...(((((((....))))))).))))))))). ( -47.60) >DroEre_CAF1 6970 110 + 1 AUCUGACUGCGUUUUCCAAUUGCCCCGGAAUUCGGCGGCCGGGAGGAGGCCGGG-GGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGCAGGCCAGGCAGCGAUCCG--- ..(((...((.((((((....((((((.....))).)))..)))))).)))))(-((((((((....)).)))))))(((((...(((((((....))))))).)))))..--- ( -51.50) >DroYak_CAF1 6541 109 + 1 AUCUGACUGCAUUUUCCAAUUGCCCUGGAAUUCGGUGGCCA-----AGCCCGGGAGGCAUCCCUUGUGGCAACGCUUGAUUGAAAUGCCUGGCAGGCCAGGCAGCGAUCCCAGC .....((((....(((((.......)))))..)))).((((-----.....((((....))))...))))...(((.(((((...(((((((....))))))).)))))..))) ( -39.60) >consensus AUCUGACUGCAUUUUCCAAUUGCCCCGGAAUUCGGUGGCCA_____AGUCCGGGAGGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGC ....((.(((......(((((..(((((.....................)))))(((((((((....)).))))))))))))...(((((((....)))))))))).))..... (-37.84 = -37.80 + -0.04)

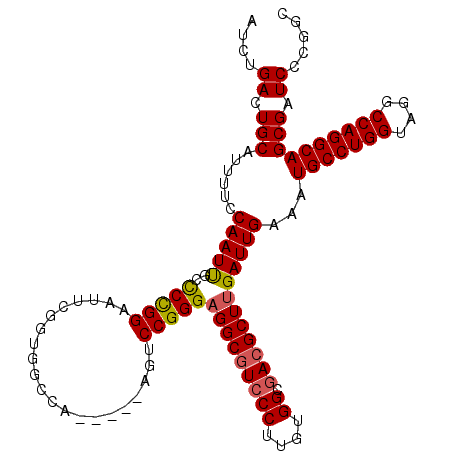
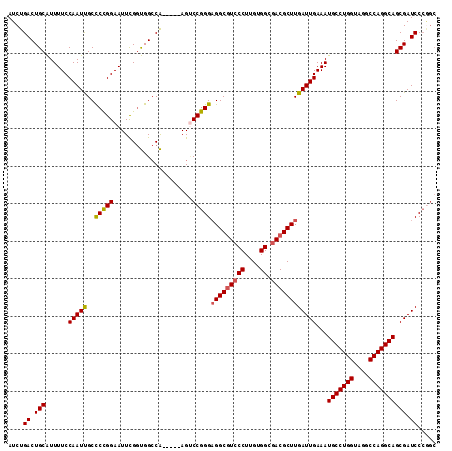
| Location | 20,490,346 – 20,490,455 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.27 |
| Mean single sequence MFE | -43.26 |
| Consensus MFE | -35.74 |
| Energy contribution | -35.74 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20490346 109 - 22407834 GCCGGGAUCGCUGCCUGGCCUACCAGGCAUUUCAAUCAAGCGUCACCACAAGGGACGCCUCCCGGACU-----UGGCCACCAAAUUCCGGAGCAAUUGGAAAAUGCAGUCAGAU .((((((....(((((((....)))))))..........(((((.((....))))))).))))))..(-----((((..((.......)).(((.((....))))).))))).. ( -40.60) >DroSec_CAF1 7394 109 - 1 GCCGGGAUCGCUGCCUGGCCUACCAGGCAUUUCAAUCAAGCGUCGCCACAAGGGACGCCUCCCGGACU-----UGGCCACCGAAUUCCAGGGCGAUUGGAAAAUGCAGUCAGAU .....(((.(((((((((....)))))))((((((((..(((((.((....)))))))..((((((.(-----(((...))))..))).))).))))))))...)).))).... ( -42.70) >DroSim_CAF1 12537 108 - 1 GCCGGGAUCGCUGCCUGGCCUACCAGGCAUUUCAAUCAAGCGUCGCCACAAGGGACGCCUCCCGGACU-----UGCCCA-CGAAUUCCGGGGCGAUUGGAAAAUGCAGUCAGAU .((((((....(((((((....)))))))..........(((((.((....))))))).))))))...-----.((((.-((.....))))))(((((.......))))).... ( -47.20) >DroEre_CAF1 6970 110 - 1 ---CGGAUCGCUGCCUGGCCUGCCAGGCAUUUCAAUCAAGCGUCGCCACAAGGGACGCC-CCCGGCCUCCUCCCGGCCGCCGAAUUCCGGGGCAAUUGGAAAACGCAGUCAGAU ---..(((.(((((((((....)))))))(((((((.....(((.((....)))))(((-(((((((.......))))).........))))).)))))))...)).))).... ( -45.40) >DroYak_CAF1 6541 109 - 1 GCUGGGAUCGCUGCCUGGCCUGCCAGGCAUUUCAAUCAAGCGUUGCCACAAGGGAUGCCUCCCGGGCU-----UGGCCACCGAAUUCCAGGGCAAUUGGAAAAUGCAGUCAGAU .(((((((((..(((.((((((..((((((..(......((...)).....)..))))))..))))))-----.)))...)).))))))).(((.((....)))))........ ( -40.40) >consensus GCCGGGAUCGCUGCCUGGCCUACCAGGCAUUUCAAUCAAGCGUCGCCACAAGGGACGCCUCCCGGACU_____UGGCCACCGAAUUCCGGGGCAAUUGGAAAAUGCAGUCAGAU .((((((....(((((((....)))))))..........(((((.((....))))))).)))))).........(((...((..((((((.....))))))..))..))).... (-35.74 = -35.74 + -0.00)

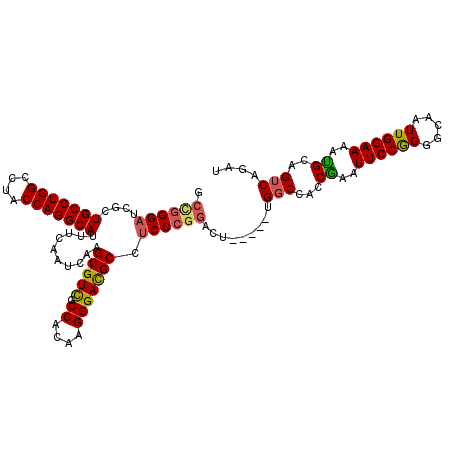
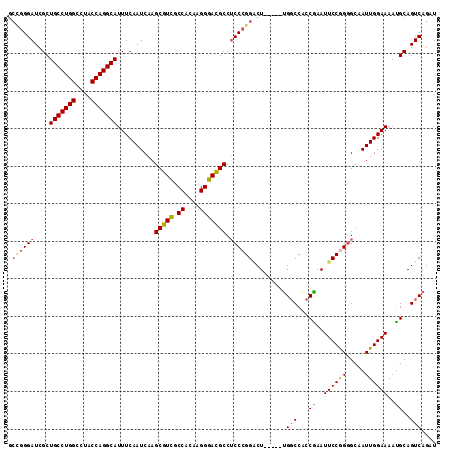
| Location | 20,490,386 – 20,490,478 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -45.54 |
| Consensus MFE | -38.76 |
| Energy contribution | -39.36 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20490386 92 + 22407834 A-----AGUCCGGGAGGCGUCCCUUGUGGUGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGCCACUC------------CCACUCGCAGCCCGGAGC .-----..((((((..(((......((((.((.(((.(((((...(((((((....))))))).)))))..)))...))------------)))).)))..)))))).. ( -44.90) >DroSec_CAF1 7434 104 + 1 A-----AGUCCGGGAGGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGCCACUCCCACUUCCACUCCCACUCGCAGCCCGGAGC .-----..((((((..(((((((....)).)))(((.(((((...(((((((....))))))).)))))..))).......................))..)))))).. ( -44.00) >DroSim_CAF1 12576 104 + 1 A-----AGUCCGGGAGGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGCCACUCCCACUUCCACUCCCACUCGCAGCCCGGAGC .-----..((((((..(((((((....)).)))(((.(((((...(((((((....))))))).)))))..))).......................))..)))))).. ( -44.00) >DroEre_CAF1 7010 95 + 1 GGGAGGAGGCCGGG-GGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGCAGGCCAGGCAGCGAUCCG-------------GCCACGCCCACUCGCAGCCCGGGGC (((.(..(((((.(-((((((((....)).)))))))(((((...(((((((....))))))).)))))))-------------))).).))).((((.....)))).. ( -51.60) >DroYak_CAF1 6581 104 + 1 A-----AGCCCGGGAGGCAUCCCUUGUGGCAACGCUUGAUUGAAAUGCCUGGCAGGCCAGGCAGCGAUCCCAGCAACUCCCACUUCCACUCCCACUCGCCGCCCGGAGC .-----.(((((((.(((.........(....)(((.(((((...(((((((....))))))).)))))..))).......................))).))))).)) ( -43.20) >consensus A_____AGUCCGGGAGGCGUCCCUUGUGGCGACGCUUGAUUGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGCCACUCCCACUUCCACUCCCACUCGCAGCCCGGAGC .........((((((((((((((....)).)))))))(((((...(((((((....))))))).)))))................................)))))... (-38.76 = -39.36 + 0.60)

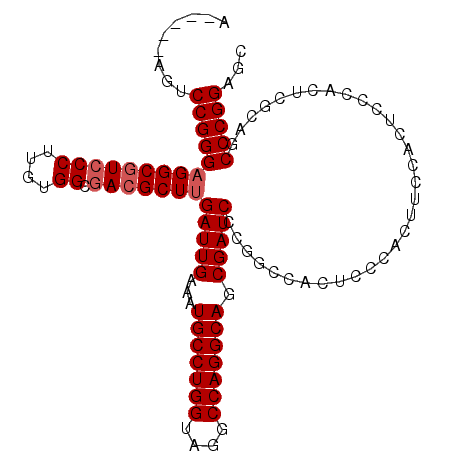
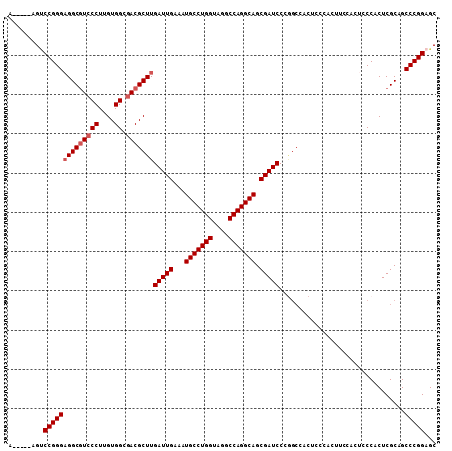
| Location | 20,490,421 – 20,490,513 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -34.01 |
| Consensus MFE | -30.59 |
| Energy contribution | -30.63 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20490421 92 + 22407834 UGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGCCACUC------------CCACUCGCAGCCCGGAGCAACUGCCAAGCUGCUCCGCACUUAAGUUGGCUUAA .....(((((((....)))))))((((....((......------------))..))))(((((((((((.((....)).)))))))..........))))... ( -34.80) >DroSec_CAF1 7469 104 + 1 UGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGCCACUCCCACUUCCACUCCCACUCGCAGCCCGGAGCAACUGCCAAGCUGCUCCGCACUUAAGUUGGCUUAA .....(((((((....)))))))((((....((.................))...))))(((((((((((.((....)).)))))))..........))))... ( -33.23) >DroSim_CAF1 12611 104 + 1 UGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGCCACUCCCACUUCCACUCCCACUCGCAGCCCGGAGCAACUGCCAAGCUGCUCCGCACUUAAGUUGGCUUAA .....(((((((....)))))))((((....((.................))...))))(((((((((((.((....)).)))))))..........))))... ( -33.23) >DroEre_CAF1 7049 91 + 1 UGAAAUGCCUGGCAGGCCAGGCAGCGAUCCG-------------GCCACGCCCACUCGCAGCCCGGGGCAACUGGCAAGCUGGUCCGCACUUAAGUUGGCUUAA .....(((((((....)))))))(((..(((-------------((...((......)).(((..((....)))))..)))))..)))...((((....)))). ( -36.80) >DroYak_CAF1 6616 104 + 1 UGAAAUGCCUGGCAGGCCAGGCAGCGAUCCCAGCAACUCCCACUUCCACUCCCACUCGCCGCCCGGAGCAACUGGCAAGCUGCUCCGCACUUAAGUUGGCUUAA .....(((((((....)))))))((.......)).......................(((...(((((((.((....)).))))))).((....)).))).... ( -32.00) >consensus UGAAAUGCCUGGUAGGCCAGGCAGCGAUCCCGGCCACUCCCACUUCCACUCCCACUCGCAGCCCGGAGCAACUGCCAAGCUGCUCCGCACUUAAGUUGGCUUAA .....(((((((....)))))))((((............................)))).((((((((((.((....)).)))))))..........))).... (-30.59 = -30.63 + 0.04)

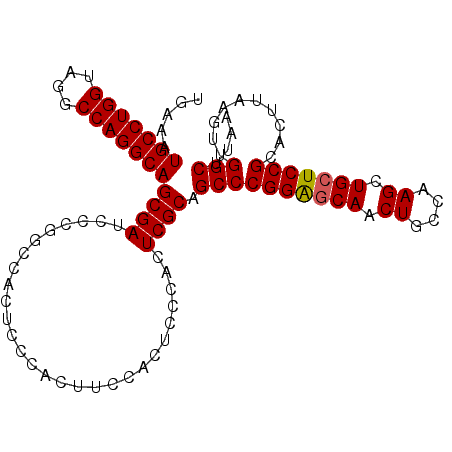
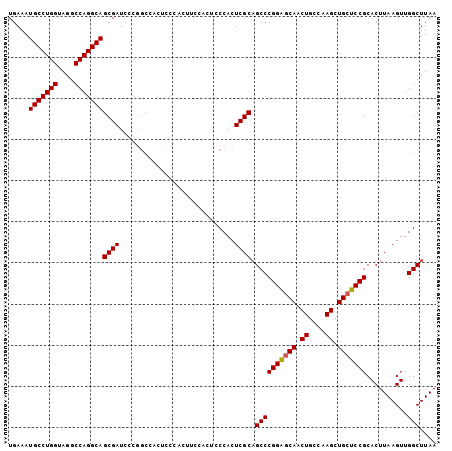
| Location | 20,490,421 – 20,490,513 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -45.32 |
| Consensus MFE | -36.94 |
| Energy contribution | -37.54 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20490421 92 - 22407834 UUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCGAGUGG------------GAGUGGCCGGGAUCGCUGCCUGGCCUACCAGGCAUUUCA ....(((.((((.(((.((((((((((....)))))))))).)))..))))((------------....(((((((.......)))))))..)).)))...... ( -46.90) >DroSec_CAF1 7469 104 - 1 UUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCGAGUGGGAGUGGAAGUGGGAGUGGCCGGGAUCGCUGCCUGGCCUACCAGGCAUUUCA .((..((.((((.(((.((((((((((....)))))))))).)))..)))).))..))((((((((...(((((((.......)))))))..))...)))))). ( -48.50) >DroSim_CAF1 12611 104 - 1 UUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCGAGUGGGAGUGGAAGUGGGAGUGGCCGGGAUCGCUGCCUGGCCUACCAGGCAUUUCA .((..((.((((.(((.((((((((((....)))))))))).)))..)))).))..))((((((((...(((((((.......)))))))..))...)))))). ( -48.50) >DroEre_CAF1 7049 91 - 1 UUAAGCCAACUUAAGUGCGGACCAGCUUGCCAGUUGCCCCGGGCUGCGAGUGGGCGUGGC-------------CGGAUCGCUGCCUGGCCUGCCAGGCAUUUCA ....(((.((((.(((.(((..(((((....)))))..))).)))..)))).((((.(((-------------(((........)))))))))).)))...... ( -39.60) >DroYak_CAF1 6616 104 - 1 UUAAGCCAACUUAAGUGCGGAGCAGCUUGCCAGUUGCUCCGGGCGGCGAGUGGGAGUGGAAGUGGGAGUUGCUGGGAUCGCUGCCUGGCCUGCCAGGCAUUUCA .....((.((((..((.((((((((((....)))))))))).))...)))).))(((((.((..(...)..))....)))))((((((....))))))...... ( -43.10) >consensus UUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCGAGUGGGAGUGGAAGUGGGAGUGGCCGGGAUCGCUGCCUGGCCUACCAGGCAUUUCA .....(((.(((.(((.((((((((((....)))))))))).)))..)))))).....................(((....(((((((....))))))).))). (-36.94 = -37.54 + 0.60)



| Location | 20,490,455 – 20,490,553 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -29.60 |
| Energy contribution | -29.84 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20490455 98 + 22407834 CACUC------------CCACUCGCAGCCCGGAGCAACUGCCAAGCUGCUCCGCACUUAAGUUGGCUUAAUUAGUUUUUGAAAGUUAUGCCAACGCCGGCGCCUGAGCCG .....------------......((((..(((((((.((....)).)))))))..))...((((((.(((((..........))))).))))))))((((......)))) ( -32.50) >DroSec_CAF1 7503 110 + 1 CACUCCCACUUCCACUCCCACUCGCAGCCCGGAGCAACUGCCAAGCUGCUCCGCACUUAAGUUGGCUUAAUUAGUUUUUGAAAGUUAUGCCAACGCCGGAGCCUGAGCCG ..............(((...(((.(.((.(((((((.((....)).))))))).......((((((.(((((..........))))).)))))))).))))...)))... ( -31.00) >DroSim_CAF1 12645 109 + 1 CACUCCCACUUCCACUCCCACUCGCAGCCCGGAGCAACUGCCAAGCUGCUCCGCACUUAAGUUGGCUUAAUUA-UUUUUGAAAGUUAUGCCAACGCCGGCGCGUGAGCCG ....................(((((.((((((((((.((....)).))))))).......((((((.(((((.-........))))).))))))...)))..)))))... ( -36.10) >DroEre_CAF1 7080 100 + 1 ----------GCCACGCCCACUCGCAGCCCGGGGCAACUGGCAAGCUGGUCCGCACUUAAGUUGGCUUAAUUAGUUUUUGAAAGUUAUGCCAACGCCGGCGCCUGAGCCG ----------((((.((((............))))...))))..(((.(..(((......((((((.(((((..........))))).))))))....)))..).))).. ( -30.60) >DroYak_CAF1 6650 110 + 1 AACUCCCACUUCCACUCCCACUCGCCGCCCGGAGCAACUGGCAAGCUGCUCCGCACUUAAGUUGGCUUAAUUAGUUUUUGAAAGUUAUGCCAACGCCGGCGCCUGAGCCG ..............(((.....((((((.(((((((.((....)).))))))).......((((((.(((((..........))))).))))))).)))))...)))... ( -33.80) >consensus CACUCCCACUUCCACUCCCACUCGCAGCCCGGAGCAACUGCCAAGCUGCUCCGCACUUAAGUUGGCUUAAUUAGUUUUUGAAAGUUAUGCCAACGCCGGCGCCUGAGCCG ....................((((..((((((((((.((....)).))))))).......((((((.(((((..........))))).))))))...)))...))))... (-29.60 = -29.84 + 0.24)



| Location | 20,490,455 – 20,490,553 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -41.03 |
| Consensus MFE | -33.88 |
| Energy contribution | -34.68 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20490455 98 - 22407834 CGGCUCAGGCGCCGGCGUUGGCAUAACUUUCAAAAACUAAUUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCGAGUGG------------GAGUG ..((((...(((..((((((((.(((..............))).))))))...(((.((((((((((....)))))))))).)))))..))).------------)))). ( -38.74) >DroSec_CAF1 7503 110 - 1 CGGCUCAGGCUCCGGCGUUGGCAUAACUUUCAAAAACUAAUUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCGAGUGGGAGUGGAAGUGGGAGUG (.((((..(((((.((((((((.(((..............))).))))))...(((.((((((((((....)))))))))).)))....)).))))).).))).)..... ( -44.24) >DroSim_CAF1 12645 109 - 1 CGGCUCACGCGCCGGCGUUGGCAUAACUUUCAAAAA-UAAUUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCGAGUGGGAGUGGAAGUGGGAGUG (.(((..(((.((.((((((((.(((.((.......-.))))).))))))...(((.((((((((((....)))))))))).)))....)).)).)))..))).)..... ( -44.70) >DroEre_CAF1 7080 100 - 1 CGGCUCAGGCGCCGGCGUUGGCAUAACUUUCAAAAACUAAUUAAGCCAACUUAAGUGCGGACCAGCUUGCCAGUUGCCCCGGGCUGCGAGUGGGCGUGGC---------- ..(((...(((((.((((((((.(((..............))).))))))...(((.(((..(((((....)))))..))).)))....)).))))))))---------- ( -34.14) >DroYak_CAF1 6650 110 - 1 CGGCUCAGGCGCCGGCGUUGGCAUAACUUUCAAAAACUAAUUAAGCCAACUUAAGUGCGGAGCAGCUUGCCAGUUGCUCCGGGCGGCGAGUGGGAGUGGAAGUGGGAGUU (.((((...(((((.(((((((.(((..............))).)))))).......((((((((((....))))))))))).))))).....)))).)........... ( -43.34) >consensus CGGCUCAGGCGCCGGCGUUGGCAUAACUUUCAAAAACUAAUUAAGCCAACUUAAGUGCGGAGCAGCUUGGCAGUUGCUCCGGGCUGCGAGUGGGAGUGGAAGUGGGAGUG ((((......))))((((((((.(((..............))).))))))...(((.((((((((((....)))))))))).)))))....................... (-33.88 = -34.68 + 0.80)

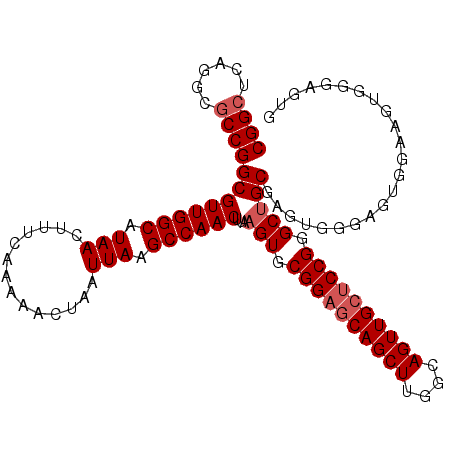
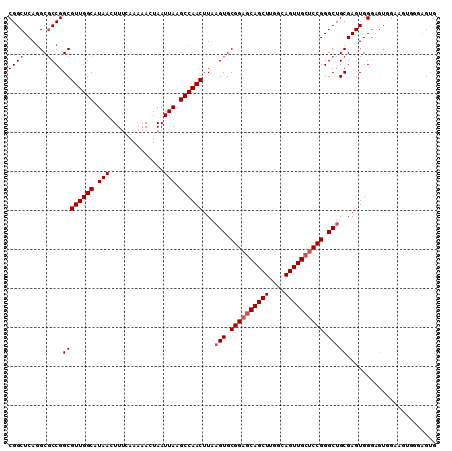
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:29 2006