| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,488,599 – 20,488,718 |
| Length | 119 |
| Max. P | 0.865129 |

| Location | 20,488,599 – 20,488,718 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.31 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20488599 119 + 22407834 UUUUGGCCAUAAUCCGCGCCGCUUUU-GUCGGCAGCUGUUUGGCUCGGCUUGGAUGCAAGUUUAAAGCUGCAACUUUUCGCCAUUGCCCGGUUUAGAUGGCACUUUAUAUUUUAUCGCCA ...((((.((((.....(((((....-).)))).(((((((((((.(((.(((.((((.((.....))))))........)))..))).)).)))))))))..........)))).)))) ( -33.60) >DroPse_CAF1 42936 98 + 1 UUUUGGCCAUAAUCUGGGCUGCCUUUUGUCGGCAGCUGUUUGGCUCGGCG-----GCAACUUUAAAGCUGCAACUUUUUGGCAUUGCC------AAGUGCCA-----------AACUCCA ...(((..........(((((((.......)))))))(((((((.(.(((-----((.........)))))......(((((...)))------))).))))-----------))).))) ( -36.70) >DroSec_CAF1 5671 119 + 1 UUUUGGCCAUAAUCCGCGCCGCUUUU-GUCGGCAGCUGUUUGGCUCGGCUUGGAUGCAAGUUUAAAGCUGCAACUUUUCGCCAUUGCCCGGCUUAAAUGGCACUUUAUAUUUUAUCGCCA ...((((.((((.....(((((....-).)))).(((((((((((.(((.(((.((((.((.....))))))........)))..))).))).))))))))..........)))).)))) ( -36.00) >DroEre_CAF1 5294 118 + 1 UUUUGGCCAUAAUCCGCGCCGCUUUUGGUCGGCAGCUGUUUGGCUCGGCU-GCAUGCAAGUUUAAAGCUGCAACUUUUCGCCAUUGC-CGGCUUAGAUGGCACUUUAUAUUCUAUCGCCA ...((((..........((((.(..(((.(((((((((.......)))))-)).((((.((.....))))))......)))))..).-))))...(((((...........))))))))) ( -36.10) >DroYak_CAF1 4811 120 + 1 UUUUGGCCAUAAUCUGCGCCGCUUUUUGUCGGCAGUUGUUUGGCGCGGCUUGGAUGCAAGUUUAAAGCUGCAACUUUUCACCAUUGCCCGGCUUAGAUGGCACUUUAUAUUUUAUCGCCA ...((((.((((...(.((((.(((..(((((((((.((..(..(((((((((((....)))).)))))))..).....)).)))).)))))..))))))).)........)))).)))) ( -33.60) >DroPer_CAF1 48726 98 + 1 UUUUGGCCAUAAUCUGGGCUGCCUUUUGUCGGCAGCUGUUUGGCUCGGCG-----GCAACUUUAAAGCUGCAACUUUUUGGCAUUGCC------AAAUGCCA-----------AACUCCA ...(((..........(((((((.......)))))))(((((((...(((-----((.........))))).....((((((...)))------))).))))-----------))).))) ( -36.40) >consensus UUUUGGCCAUAAUCCGCGCCGCUUUUUGUCGGCAGCUGUUUGGCUCGGCU_GGAUGCAAGUUUAAAGCUGCAACUUUUCGCCAUUGCCCGGCUUAAAUGGCACUUUAUAUUUUAUCGCCA ....(((((........(((((.....).)))).......))))).(((......(((..........)))........((((((..........))))))...............))). (-22.70 = -22.31 + -0.39)


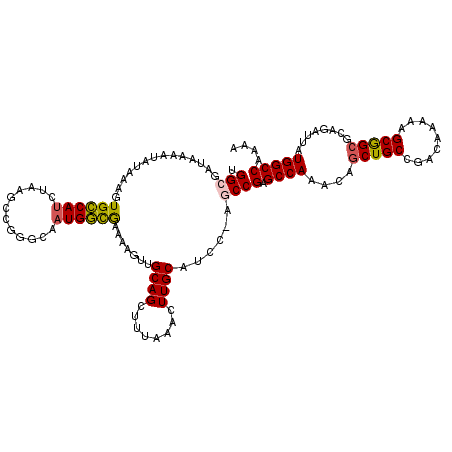
| Location | 20,488,599 – 20,488,718 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -21.92 |
| Energy contribution | -21.57 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20488599 119 - 22407834 UGGCGAUAAAAUAUAAAGUGCCAUCUAAACCGGGCAAUGGCGAAAAGUUGCAGCUUUAAACUUGCAUCCAAGCCGAGCCAAACAGCUGCCGAC-AAAAGCGGCGCGGAUUAUGGCCAAAA ((((.((((.......((......))...((((((..((((.......(((((........))))).....)))).))).....(((((....-....))))).))).)))).))))... ( -33.40) >DroPse_CAF1 42936 98 - 1 UGGAGUU-----------UGGCACUU------GGCAAUGCCAAAAAGUUGCAGCUUUAAAGUUGC-----CGCCGAGCCAAACAGCUGCCGACAAAAGGCAGCCCAGAUUAUGGCCAAAA (((.(((-----------((((..((------(((...)))))...((.((((((....))))))-----.))...))))))).((((((.......))))))(((.....))))))... ( -39.40) >DroSec_CAF1 5671 119 - 1 UGGCGAUAAAAUAUAAAGUGCCAUUUAAGCCGGGCAAUGGCGAAAAGUUGCAGCUUUAAACUUGCAUCCAAGCCGAGCCAAACAGCUGCCGAC-AAAAGCGGCGCGGAUUAUGGCCAAAA ((((.((((........(((((......((((.....)))).....(((((((((.....(((((......).))))......))))).))))-......)))))...)))).))))... ( -34.80) >DroEre_CAF1 5294 118 - 1 UGGCGAUAGAAUAUAAAGUGCCAUCUAAGCCG-GCAAUGGCGAAAAGUUGCAGCUUUAAACUUGCAUGC-AGCCGAGCCAAACAGCUGCCGACCAAAAGCGGCGCGGAUUAUGGCCAAAA ((((.((((.........((((.........)-))).((((.....((((((((.........)).)))-)))...))))....((.((((........))))))...)))).))))... ( -37.00) >DroYak_CAF1 4811 120 - 1 UGGCGAUAAAAUAUAAAGUGCCAUCUAAGCCGGGCAAUGGUGAAAAGUUGCAGCUUUAAACUUGCAUCCAAGCCGCGCCAAACAACUGCCGACAAAAAGCGGCGCAGAUUAUGGCCAAAA ((((.((((........((((.....((((...(((((........))))).)))).......))))....(((((......................))))).....)))).))))... ( -29.55) >DroPer_CAF1 48726 98 - 1 UGGAGUU-----------UGGCAUUU------GGCAAUGCCAAAAAGUUGCAGCUUUAAAGUUGC-----CGCCGAGCCAAACAGCUGCCGACAAAAGGCAGCCCAGAUUAUGGCCAAAA (((.(((-----------((((.(((------(((...))))))..((.((((((....))))))-----.))...))))))).((((((.......))))))(((.....))))))... ( -40.50) >consensus UGGCGAUAAAAUAUAAAGUGCCAUCUAAGCCGGGCAAUGGCGAAAAGUUGCAGCUUUAAACUUGCAUCC_AGCCGAGCCAAACAGCUGCCGACAAAAAGCGGCGCAGAUUAUGGCCAAAA .(((..............((((((............)))))).......((((........))))......)))(.((((....(((((.........)))))........))))).... (-21.92 = -21.57 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:23 2006