
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,487,801 – 20,487,896 |
| Length | 95 |
| Max. P | 0.789330 |

| Location | 20,487,801 – 20,487,896 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -20.14 |
| Energy contribution | -19.83 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20487801 95 + 22407834 ACGGCAAAGCAAA---------UCGGGCAACUGGGGCAAGUGCAAAGUGUAAUUAUGUGGUCACUUGGUCAGCCGUAAACUC--CGAUUUGUUUGCCCGUGUGU--------------GU ((((..(((((((---------(((((....((.(((...(((.....)))......((..(....)..))))).))...))--))))))))))..))))....--------------.. ( -29.90) >DroPse_CAF1 41705 120 + 1 ACGGAGGGGCGGAGUGGGGGGCACUGGGAACUGGGGCAAGUGCAAAGUGUAAUUAUGUGGUCGCUUGGCCAGCCCUAAACUACCAGAUUUGUUUGCCCGAGUGCUCGAAGAGCUUGUUGU ......(((((((((..(((((.(.(....).)((.(((((((...(((....)))...).)))))).)).)))))..))).))(((....)))))))....((((...))))....... ( -39.30) >DroSec_CAF1 4881 94 + 1 ACGCCAAAGCAAA---------UCGGGCAACUGGGGCAAGUGCAAAGUGUAAUUAUGUGGUCACUUGGUCAGCCGUAAACUC--CGAUUUGUUUGCC-GUGUGU--------------GU ((((..(((((((---------(((((....((.(((...(((.....)))......((..(....)..))))).))...))--))))))))))...-))))..--------------.. ( -28.00) >DroSim_CAF1 10000 94 + 1 ACGGCAAAGCAAA---------UCGGGCAACUGGGGCAAGUGCAAAGUGUAAUUAUGUGGUCACUUGGUCAGCCGUAAACUC--CGAUUUGUUUGCC-GUGUGU--------------GU ((((((((.((((---------(((((....((.(((...(((.....)))......((..(....)..))))).))...))--)))))))))))))-))....--------------.. ( -33.40) >DroEre_CAF1 4565 87 + 1 --GGCAAAGCAAA---------UCGUGCAACUGGGGCAAGUGCAAAGUGUAAUUAUGUGGUCACUUGGUCAGCCGUAAAUUC--CGAUUUGUUUGCC-A-------------------GU --((((((.((((---------((((((..(((.(.(((((((...(((....)))...).)))))).))))..))).....--)))))))))))))-.-------------------.. ( -27.30) >DroYak_CAF1 4051 87 + 1 --GGCAAAGCAAA---------UCGGGCAACUGGGGCAAGUGCAAAGUGUAAUUAUGUGGUCACUUGGUCAGCCGUAAACUC--CGAUUUGUUUGCC-A-------------------GU --((((((.((((---------(((((....((.(((...(((.....)))......((..(....)..))))).))...))--)))))))))))))-.-------------------.. ( -30.30) >consensus ACGGCAAAGCAAA_________UCGGGCAACUGGGGCAAGUGCAAAGUGUAAUUAUGUGGUCACUUGGUCAGCCGUAAACUC__CGAUUUGUUUGCC_GUGUGU______________GU ..((((((.((((..........(((....)))((.(((((((...(((....)))...).)))))).)).................))))))))))....................... (-20.14 = -19.83 + -0.30)

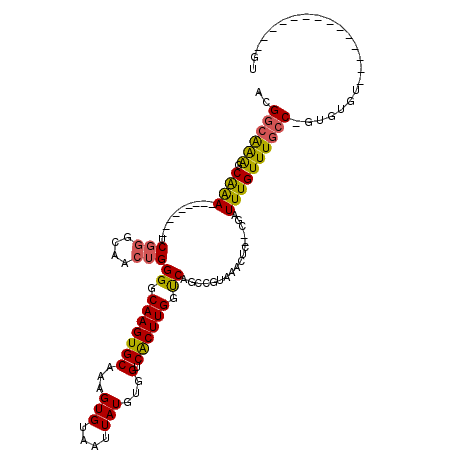
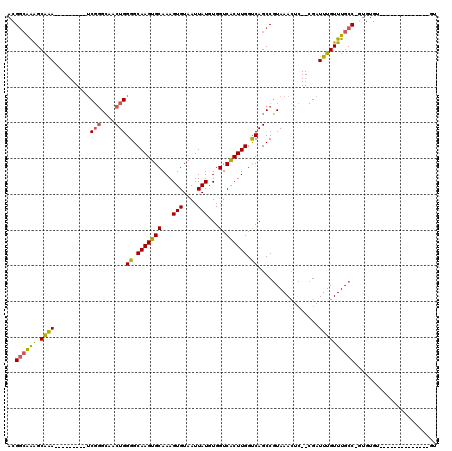
| Location | 20,487,801 – 20,487,896 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -14.09 |
| Energy contribution | -15.15 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20487801 95 - 22407834 AC--------------ACACACGGGCAAACAAAUCG--GAGUUUACGGCUGACCAAGUGACCACAUAAUUACACUUUGCACUUGCCCCAGUUGCCCGA---------UUUGCUUUGCCGU ..--------------.......(((((((((((((--(.((....(((((..((((((...................))))))...)))))))))))---------)))).)))))).. ( -29.61) >DroPse_CAF1 41705 120 - 1 ACAACAAGCUCUUCGAGCACUCGGGCAAACAAAUCUGGUAGUUUAGGGCUGGCCAAGCGACCACAUAAUUACACUUUGCACUUGCCCCAGUUCCCAGUGCCCCCCACUCCGCCCCUCCGU .......((((...))))....(((((.......((((....((.((((..((.(((.(............).))).))....)))).))...))))))))).................. ( -25.41) >DroSec_CAF1 4881 94 - 1 AC--------------ACACAC-GGCAAACAAAUCG--GAGUUUACGGCUGACCAAGUGACCACAUAAUUACACUUUGCACUUGCCCCAGUUGCCCGA---------UUUGCUUUGGCGU ..--------------....((-(.(((((((((((--(.((....(((((..((((((...................))))))...)))))))))))---------)))).)))).))) ( -27.71) >DroSim_CAF1 10000 94 - 1 AC--------------ACACAC-GGCAAACAAAUCG--GAGUUUACGGCUGACCAAGUGACCACAUAAUUACACUUUGCACUUGCCCCAGUUGCCCGA---------UUUGCUUUGCCGU ..--------------....((-(((((((((((((--(.((....(((((..((((((...................))))))...)))))))))))---------)))).)))))))) ( -32.31) >DroEre_CAF1 4565 87 - 1 AC-------------------U-GGCAAACAAAUCG--GAAUUUACGGCUGACCAAGUGACCACAUAAUUACACUUUGCACUUGCCCCAGUUGCACGA---------UUUGCUUUGCC-- ..-------------------.-(((((((((((((--.......((((((..((((((...................))))))...))))))..)))---------)))).))))))-- ( -24.81) >DroYak_CAF1 4051 87 - 1 AC-------------------U-GGCAAACAAAUCG--GAGUUUACGGCUGACCAAGUGACCACAUAAUUACACUUUGCACUUGCCCCAGUUGCCCGA---------UUUGCUUUGCC-- ..-------------------.-(((((((((((((--(.((....(((((..((((((...................))))))...)))))))))))---------)))).))))))-- ( -27.91) >consensus AC______________ACACAC_GGCAAACAAAUCG__GAGUUUACGGCUGACCAAGUGACCACAUAAUUACACUUUGCACUUGCCCCAGUUGCCCGA_________UUUGCUUUGCCGU .......................((((((((((............((((((..((((((...................))))))...))))))..............)))).)))))).. (-14.09 = -15.15 + 1.06)

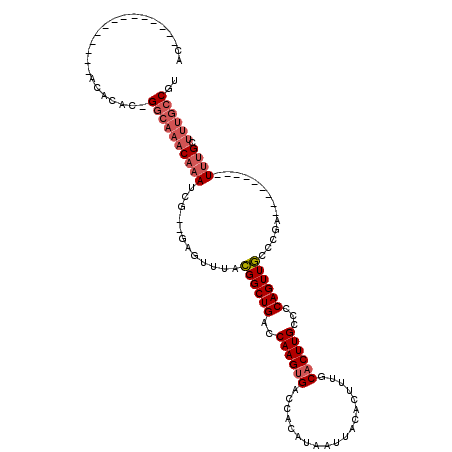
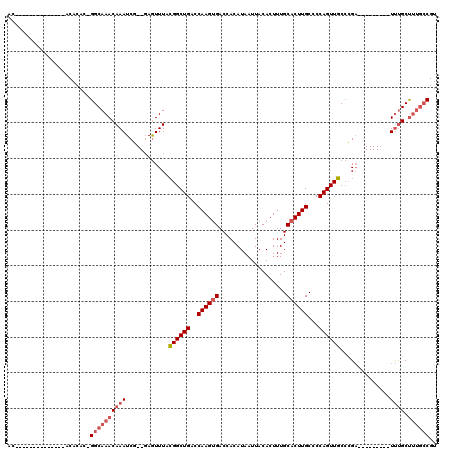
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:21 2006