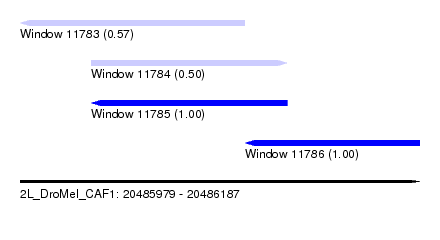
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,485,979 – 20,486,187 |
| Length | 208 |
| Max. P | 0.998826 |

| Location | 20,485,979 – 20,486,096 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -21.06 |
| Energy contribution | -23.13 |
| Covariance contribution | 2.07 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20485979 117 - 22407834 GGCUGCACUUGUCGUGCGGCUUGAGUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAGAGC--GGAAUGCUUUUGGGAGGACCGGUGGAUGGGUGAA ((((((((.....))))))))....((((((....(((((((((((.........)))))))(((.(((((((((.((.....--.)).)))))))))...))).))))..)))).)). ( -36.00) >DroPse_CAF1 40050 101 - 1 --------------UCCGGCUGGAGUCCCCCAUGAUAUCGCACAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAGAGAAG-GAGUCCUGCAG-GAGGAG-GGCGGAG-GGCGGA --------------((((.((...((((.((.(((((.((((((((.........)))))))).)))))................-...(((....)-)))).)-)))...)-).)))) ( -33.90) >DroSec_CAF1 3092 106 - 1 GGCUGCACUUGUCGUGCGGCUUGAGUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAGAGC--GGAAUGCUGUUGGG-----------AUGGGUGGA ((((((((.....))))))))...(((((...(((((.((((((((.........)))))))).)))))...........(((--((....))))))))-----------))....... ( -32.90) >DroSim_CAF1 8458 106 - 1 GGCUGCACUUGUCGUGCGGCUUGAGUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAGAGC--GGAAUGCUGUUGGG-----------AUGGGUGGA ((((((((.....))))))))...(((((...(((((.((((((((.........)))))))).)))))...........(((--((....))))))))-----------))....... ( -32.90) >DroEre_CAF1 2980 92 - 1 GGCUGCACUUGUUGUGCGGCAGGACUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAGAGC--GGAAUGCUG------------------------- .(((((((.....)))))))((.(.(((....(((((.((((((((.........)))))))).)))))...((.......))--))).).)).------------------------- ( -29.20) >DroYak_CAF1 2301 117 - 1 GGCUGCACUUGUUGUGCGGCUUGACUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAGAGC--GGAAUGCUGGUGGGAGGACUGGUGGAUAGGUGGA ((((((((.....))))))))...((((((..(((((.((((((((.........)))))))).)))))..((((.((.....--.)).)))).).)))))..(((.....)))..... ( -35.40) >consensus GGCUGCACUUGUCGUGCGGCUUGAGUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAGAGC__GGAAUGCUGUUGGG___________AUGGGUGGA .(((((((.....))))))).......((((.(((((.((((((((.........)))))))).)))))..((((.((........)).))))..)))).................... (-21.06 = -23.13 + 2.07)

| Location | 20,486,016 – 20,486,118 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20486016 102 + 22407834 CUAAUCGCUUCUAAUAACGCACAAAAUUGAAAAACUUAUGCGAUAUCAUUGGGGACUCAAGCCGCACGACAAGUGCAGCCGCCCUGCGGGGGGAUUCCC---------CAA ...(((((...........((......))..........)))))....(((((((.((...((((((.....))))..((((...)))))).)).))))---------))) ( -30.50) >DroSec_CAF1 3118 102 + 1 CUAAUCGCUUCUAAUAACGCACAAAAUUGAAAAACUUAUGCGAUAUCAUUGGGGACUCAAGCCGCACGACAAGUGCAGCCGCCCUGCGGGGGGAUUCCC---------CAA ...(((((...........((......))..........)))))....(((((((.((...((((((.....))))..((((...)))))).)).))))---------))) ( -30.50) >DroSim_CAF1 8484 102 + 1 CUAAUCGCUUCUAAUAACGCACAAAAUUGAAAAACUUAUGCGAUAUCAUUGGGGACUCAAGCCGCACGACAAGUGCAGCCGCCCUGCGGGGGGAUUCCC---------CAA ...(((((...........((......))..........)))))....(((((((.((...((((((.....))))..((((...)))))).)).))))---------))) ( -30.50) >DroEre_CAF1 2992 101 + 1 CUAAUCGCUUCUAAUAACGCACAAAAUUGAAAAACUUAUGCGAUAUCAUUGGGGAGUCCUGCCGCACAACAAGUGCAGCCGCCCUGCGGG-GGAUUCCC---------CAA ...(((((...........((......))..........)))))....((((((((((((...((((.....))))..((((...)))))-))))))))---------))) ( -36.10) >DroYak_CAF1 2338 111 + 1 CUAAUCGCUUCUAAUAACGCACAAAAUUGAAAAACUUAUGCGAUAUCAUUGGGGAGUCAAGCCGCACAACAAGUGCAGCCGCCCUGCGGGGGGAUUCCCCAGGAAUCCCAA ...(((((...........((......))..........))))).((.((((((((((...((((((.....))))..((((...)))))).))))))))))))....... ( -34.70) >consensus CUAAUCGCUUCUAAUAACGCACAAAAUUGAAAAACUUAUGCGAUAUCAUUGGGGACUCAAGCCGCACGACAAGUGCAGCCGCCCUGCGGGGGGAUUCCC_________CAA ...(((((...........((......))..........))))).......((((.((..((.((((.....)))).))(.(((...))).))).))))............ (-23.10 = -23.10 + -0.00)


| Location | 20,486,016 – 20,486,118 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.32 |
| Mean single sequence MFE | -40.26 |
| Consensus MFE | -32.94 |
| Energy contribution | -33.34 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20486016 102 - 22407834 UUG---------GGGAAUCCCCCCGCAGGGCGGCUGCACUUGUCGUGCGGCUUGAGUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAG ..(---------(((....))))(((.((((((((((((.....))))))))...))))....(((((.((((((((.........)))))))).)))))...)))..... ( -39.00) >DroSec_CAF1 3118 102 - 1 UUG---------GGGAAUCCCCCCGCAGGGCGGCUGCACUUGUCGUGCGGCUUGAGUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAG ..(---------(((....))))(((.((((((((((((.....))))))))...))))....(((((.((((((((.........)))))))).)))))...)))..... ( -39.00) >DroSim_CAF1 8484 102 - 1 UUG---------GGGAAUCCCCCCGCAGGGCGGCUGCACUUGUCGUGCGGCUUGAGUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAG ..(---------(((....))))(((.((((((((((((.....))))))))...))))....(((((.((((((((.........)))))))).)))))...)))..... ( -39.00) >DroEre_CAF1 2992 101 - 1 UUG---------GGGAAUCC-CCCGCAGGGCGGCUGCACUUGUUGUGCGGCAGGACUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAG (((---------((((.(((-..(((...)))(((((((.....))))))).))).)))))))(((((.((((((((.........)))))))).)))))........... ( -41.00) >DroYak_CAF1 2338 111 - 1 UUGGGAUUCCUGGGGAAUCCCCCCGCAGGGCGGCUGCACUUGUUGUGCGGCUUGACUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAG ..((((((((...))))))))..(((.(((.((((((((.....))))))))......)))..(((((.((((((((.........)))))))).)))))...)))..... ( -43.30) >consensus UUG_________GGGAAUCCCCCCGCAGGGCGGCUGCACUUGUCGUGCGGCUUGAGUCCCCAAUGAUAUCGCAUAAGUUUUUCAAUUUUGUGCGUUAUUAGAAGCGAUUAG ............(((......)))((.((((.(((((((.....)))))))....))))....(((((.((((((((.........)))))))).)))))...))...... (-32.94 = -33.34 + 0.40)

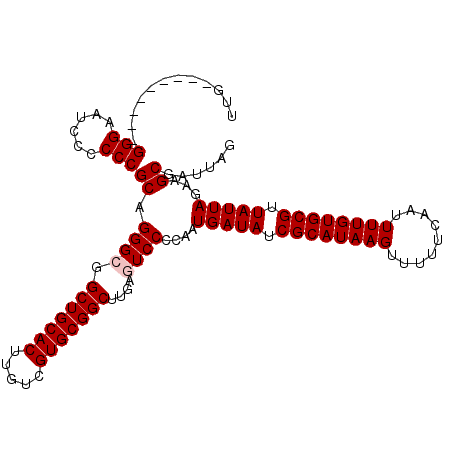

| Location | 20,486,096 – 20,486,187 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -31.04 |
| Energy contribution | -31.24 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20486096 91 - 22407834 CUCGUCCUGUUUACUUUGCUGGCCCUGCCUCAUGUGCUUAAAGUUGUCACAAGUUGUCGGGUCCAGUUCUUG---------GGGAAUCCCCCCGCAGGGC ...(((((((.......((((((((.((..(.(((((........).)))).)..)).))).)))))....(---------(((.....))))))))))) ( -35.60) >DroSec_CAF1 3198 91 - 1 CUCGUCCUGUUUACUUUGCUGGCCCUGCCUCAUGUGCUUAAAGUUGUCACAAGUUGUCGGGUCCAGUUCUUG---------GGGAAUCCCCCCGCAGGGC ...(((((((.......((((((((.((..(.(((((........).)))).)..)).))).)))))....(---------(((.....))))))))))) ( -35.60) >DroSim_CAF1 8564 91 - 1 CUCGUCCUGUUUACUUUGCUGGCCCUGCCUCAUGUGCUUAAAGUUGUCACAAGUUGUCGGGUCCAGUUCUUG---------GGGAAUCCCCCCGCAGGGC ...(((((((.......((((((((.((..(.(((((........).)))).)..)).))).)))))....(---------(((.....))))))))))) ( -35.60) >DroEre_CAF1 3072 90 - 1 CUCGUCCUGUUUACUUUGCGGGCCCUGCCUCAUGUGCUUAAAGUUGUCACAAGUUGUCGGGUCCAGUUCUUG---------GGGAAUCC-CCCGCAGGGC ...(((((((.........((((((.((..(.(((((........).)))).)..)).)))))).......(---------(((....)-)))))))))) ( -35.50) >DroYak_CAF1 2418 100 - 1 CUCGUCCUGUUGACUUUGCUGGCCCUGCCCCAUGUGCUUAAAGUUGUCACAAGUUGUGGGGUCCAGUUCUUGGGAUUCCUGGGGAAUCCCCCCGCAGGGC ...(((((((.(.....((((((((..(..(.(((((........).)))).)..)..))).)))))....((((((((...)))))))).).))))))) ( -41.80) >consensus CUCGUCCUGUUUACUUUGCUGGCCCUGCCUCAUGUGCUUAAAGUUGUCACAAGUUGUCGGGUCCAGUUCUUG_________GGGAAUCCCCCCGCAGGGC ...(((((((.......((((((((..(..(.(((((........).)))).)..)..))).)))))..............(((......)))))))))) (-31.04 = -31.24 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:17 2006