
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,483,733 – 20,483,904 |
| Length | 171 |
| Max. P | 0.929058 |

| Location | 20,483,733 – 20,483,833 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.97 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.82 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20483733 100 + 22407834 CAGCAAACAGCGCCGGGAAAAUC----------------GGAAAACCUCAGUGCAAACCGAGGGACGAGUGACAGUGUGUUGUUGUCCGCUCUGCGUUUGCACAUCAAAUCAAAUG ..((.....)).((((.....))----------------)).........((((((((((((((((.((..((.....))..)))))).))).).))))))))............. ( -30.50) >DroSec_CAF1 885 100 + 1 CAGCAAACAGCGCCGGGAAAAUC----------------GGAAAACCUCAGUGCAAACCGAGGGACGAGUGACAGCGUGUUGUUGUCCGCUCUGCGUUUGCACAUCAAAUCAGAUG ..((((((.((.((((.....))----------------))....((((.((....)).))))...((((((((((.....))))).))))).)))))))).((((......)))) ( -34.70) >DroSim_CAF1 6267 100 + 1 CAGCAAACAGCGCCGGGAAAAUC----------------GGAAAACCUCAGUGCAAACCGAGGGACGAGUGACAGCGUGUUGUUGUCCGCUCUGCGUUUGCACAUCAAAUCAGAUG ..((((((.((.((((.....))----------------))....((((.((....)).))))...((((((((((.....))))).))))).)))))))).((((......)))) ( -34.70) >DroEre_CAF1 825 116 + 1 CAGCAAACAGCGUCGGGAAAAUCGUCGGGAAACUCGCUGGGAAAACCUCAGUGCCAACCGAGGGACGACUGACAGCGUGUUGUUGACCGCCCUGCGUUUGCACAUCAAAUCAAAUG ..((((((.(((((((.....((.((((......(((((((.....)))))))....)))).))....))))).(((.(((...))))))...))))))))............... ( -37.50) >DroYak_CAF1 100 100 + 1 CAGCAAACAGCGUCGGAAAAAUC----------------GGAAAACCUCAGUGAAAACCAAGGGUCGAGUGACAGCGCGUUGUUGACCGACCUGCGCUUGCACAUCAAAUCAAAUG ..((((.(((.(((((.....((----------------((....(((..((....))..))).))))..(((((....)))))..))))))))...))))............... ( -26.90) >consensus CAGCAAACAGCGCCGGGAAAAUC________________GGAAAACCUCAGUGCAAACCGAGGGACGAGUGACAGCGUGUUGUUGUCCGCUCUGCGUUUGCACAUCAAAUCAAAUG ..((((...((((.(((.............................(((.((....)).)))((((((.((((.....)))))))))).))).))))))))............... (-23.80 = -23.82 + 0.02)

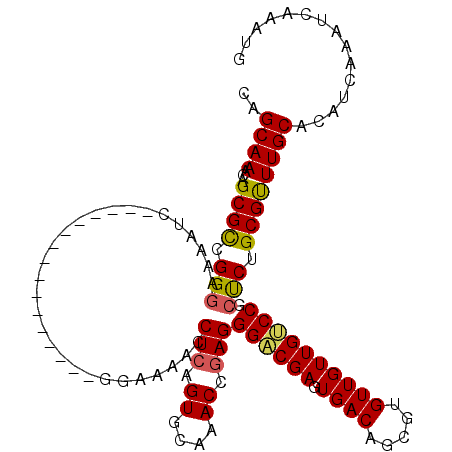

| Location | 20,483,793 – 20,483,904 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.63 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -10.60 |
| Energy contribution | -11.74 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
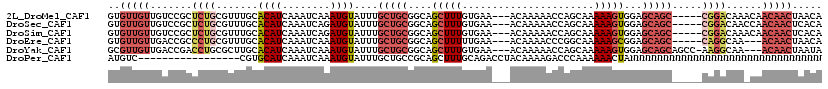
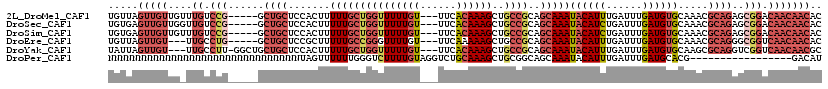
>2L_DroMel_CAF1 20483793 111 + 22407834 GUGUUGUUGUCCGCUCUGCGUUUGCACAUCAAAUCAAAUGUAUUUGCUGCGGCAGCUUUGUGAA---ACAAAAACCAGCAAAAAGUGGAGCAGC-----CGGACAAACAACAACUAACA .((((((((((((..((((.((..(((((........)))).(((((((.(.....((((....---.))))..))))))))..)..)))))).-----))))).)))))))....... ( -34.70) >DroSec_CAF1 945 111 + 1 GUGUUGUUGUCCGCUCUGCGUUUGCACAUCAAAUCAGAUGUAUUUGCUGCGGCAGCUUUGUGAA---ACAAAAACCAGCAAAAAGUGGAGCAGC-----CGGACAACCAACAACUCACA .((((((((((((..((((.((..((((((......))))).(((((((.(.....((((....---.))))..))))))))..)..)))))).-----))))))).)))))....... ( -38.60) >DroSim_CAF1 6327 111 + 1 GUGUUGUUGUCCGCUCUGCGUUUGCACAUCAAAUCAGAUGUAUUUGCUGCGGCAGCUUUGUGAA---ACAAAAACCAGCAAAAAGUGGAGCAGC-----CGGACAAACAACAACUCACA .((((((((((((..((((.((..((((((......))))).(((((((.(.....((((....---.))))..))))))))..)..)))))).-----))))).)))))))....... ( -38.60) >DroEre_CAF1 901 108 + 1 GUGUUGUUGACCGCCCUGCGUUUGCACAUCAAAUCAAAUGUAUUUGCUGCGGCAGCUUUUUGAA---ACAAAACCCGGCAAAAAGCGGAGCAGC-----CAGGCAA---ACAACUAACA ..((((((....(((.((((((((..........))))))))...(((((....((((((((..---.(.......).))))))))...)))))-----..))).)---)))))..... ( -31.70) >DroYak_CAF1 160 112 + 1 GCGUUGUUGACCGACCUGCGCUUGCACAUCAAAUCAAAUGUAUUUGCUGCGGCAGCUUUGUGAA---ACAAAAACCAGCAAAAAGUGGAGCAGCAGCC-AAGGCAA---ACAACUAAUA ..((((((..((...(((((((..(((((........)))).(((((((.(.....((((....---.))))..))))))))..)..).)).))))..-..))..)---)))))..... ( -28.80) >DroPer_CAF1 43244 102 + 1 AUGUC-----------------CGUGCAUCAAAUCAAAUGUAUUUGCUGCCGCAGCUUUGCAGACCUACAAAAGACCCAAAAAACUANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ..(((-----------------.(.(((.((((((....).))))).))))((......)).)))...................................................... ( -9.30) >consensus GUGUUGUUGUCCGCUCUGCGUUUGCACAUCAAAUCAAAUGUAUUUGCUGCGGCAGCUUUGUGAA___ACAAAAACCAGCAAAAAGUGGAGCAGC_____CGGACAA___ACAACUAACA ..(((((.......((((.......((((........))))....(((((....(((((......................)))))...))))).....))))......)))))..... (-10.60 = -11.74 + 1.14)
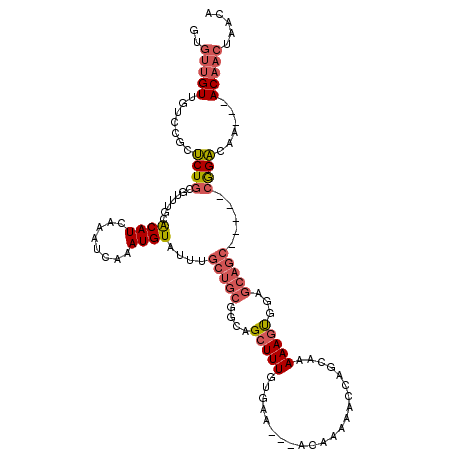
| Location | 20,483,793 – 20,483,904 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.63 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -14.88 |
| Energy contribution | -16.45 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
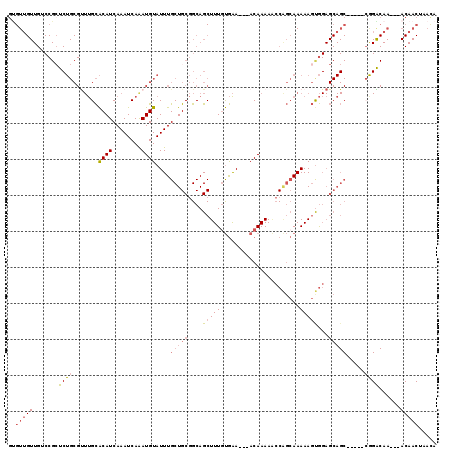
>2L_DroMel_CAF1 20483793 111 - 22407834 UGUUAGUUGUUGUUUGUCCG-----GCUGCUCCACUUUUUGCUGGUUUUUGU---UUCACAAAGCUGCCGCAGCAAAUACAUUUGAUUUGAUGUGCAAACGCAGAGCGGACAACAACAC .......(((((((.(((((-----.((((.......((((((((((((((.---....)))))..)))..))))))((((((......)))))).....))))..)))))))))))). ( -35.50) >DroSec_CAF1 945 111 - 1 UGUGAGUUGUUGGUUGUCCG-----GCUGCUCCACUUUUUGCUGGUUUUUGU---UUCACAAAGCUGCCGCAGCAAAUACAUCUGAUUUGAUGUGCAAACGCAGAGCGGACAACAACAC .......(((((.(((((((-----.((((.......((((((((((((((.---....)))))..)))..))))))((((((......)))))).....))))..)))))))))))). ( -37.80) >DroSim_CAF1 6327 111 - 1 UGUGAGUUGUUGUUUGUCCG-----GCUGCUCCACUUUUUGCUGGUUUUUGU---UUCACAAAGCUGCCGCAGCAAAUACAUCUGAUUUGAUGUGCAAACGCAGAGCGGACAACAACAC .......(((((((.(((((-----.((((.......((((((((((((((.---....)))))..)))..))))))((((((......)))))).....))))..)))))))))))). ( -37.80) >DroEre_CAF1 901 108 - 1 UGUUAGUUGU---UUGCCUG-----GCUGCUCCGCUUUUUGCCGGGUUUUGU---UUCAAAAAGCUGCCGCAGCAAAUACAUUUGAUUUGAUGUGCAAACGCAGGGCGGUCAACAACAC .....(((((---.....((-----(((((((.((..(((((.((((...((---((....)))).))).).)))))((((((......)))))).....)).)))))))))))))).. ( -33.30) >DroYak_CAF1 160 112 - 1 UAUUAGUUGU---UUGCCUU-GGCUGCUGCUCCACUUUUUGCUGGUUUUUGU---UUCACAAAGCUGCCGCAGCAAAUACAUUUGAUUUGAUGUGCAAGCGCAGGUCGGUCAACAACGC .....(((((---(.(((..-.(((((.((.((((.....).))).(((((.---....)))))..)).)))))...............(((.(((....))).)))))).)))))).. ( -32.70) >DroPer_CAF1 43244 102 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUAGUUUUUUGGGUCUUUUGUAGGUCUGCAAAGCUGCGGCAGCAAAUACAUUUGAUUUGAUGCACG-----------------GACAU ................................(((((((.(((..((.....))..))).)))))))(((((.(((((.(....)))))).))).))-----------------..... ( -17.10) >consensus UGUUAGUUGU___UUGUCCG_____GCUGCUCCACUUUUUGCUGGUUUUUGU___UUCACAAAGCUGCCGCAGCAAAUACAUUUGAUUUGAUGUGCAAACGCAGAGCGGACAACAACAC .....(((((....((.(((......((((.......(((((((((((((((......))))))..))))..)))))((((((......)))))).....))))..))).))))))).. (-14.88 = -16.45 + 1.57)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:14 2006