
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,482,620 – 20,482,859 |
| Length | 239 |
| Max. P | 0.682920 |

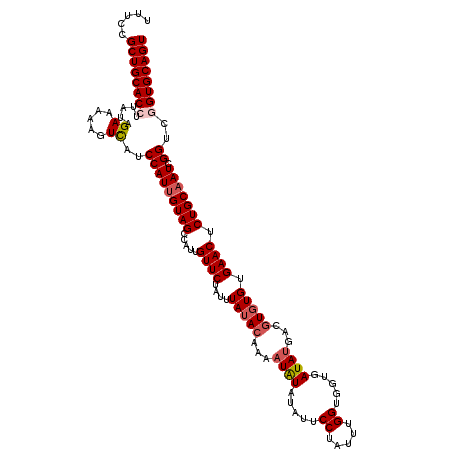
| Location | 20,482,620 – 20,482,740 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -24.64 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20482620 120 - 22407834 UUUCCGCUGCACCUUAUAGAAAAAGUUAUCCAUAGUAGACAUUGUUCUAUUUAUACAAAAUAUAUAUUCCGAUUUGGUGGUGAUAUGAUGUGUGUGAACUCUGCAAUCGGUCGGUGCAGU .....((((((((((((.((........)).))))..((((((((....((((((((...(((((...(((......)))..)))))...))))))))....)))))..))))))))))) ( -30.90) >DroSec_CAF1 35543 120 - 1 UUUCCGCUGCACCUUAUAGAAAAAGUCAUCCAUUGUAGCCAUGGUUCUAUUUAUACAAAAUAUAUAUUCCUGUUUGGUGGUGAUAUGACGUGUGUGAACUCUGCAAUCGGUCGGUGCAGU .....((((((((.....((.....))..(((((((((....(((((....(((((...((((.....((.....)).....))))...))))).)))))))))))).))..)))))))) ( -32.10) >DroSim_CAF1 39366 120 - 1 UUUCCGCUGCACCUUAUAGAAAAAGUCAUCCAUUGUAGCCAUGGUUCUAUUUAUACAAAAUAUAUAUUCCUGUUUGGUGGUGAUAUGACGUGUGUGAACUCUGCAAUCGGUCGGUGCAGU .....((((((((.....((.....))..(((((((((....(((((....(((((...((((.....((.....)).....))))...))))).)))))))))))).))..)))))))) ( -32.10) >DroEre_CAF1 37942 119 - 1 UUUCCGCUGCACCUUGUGGAAAUAAUCAUCCAUUGUAGCCAUUGUUCUAUUUAUACA-AAUGUAUAUUCCUAUUCGGUGGUGAUAAGUCGUGUGUGAACUCUGCAAUCGGUCAGUGCAGU .....(((((((...(((((........)))))....((((((((....((((((((-....(((...((........))..))).....))))))))....))))).)))..))))))) ( -30.60) >DroYak_CAF1 35294 119 - 1 UUUCCGCUGCACCUUAUGGAAGAAAUCAUCCAUUGUAGCCAUUGUUCUAUUUAAAGA-AAUAUAUAUUCCAAUUUGGUGGUGAUAUGACGUGUGUGAACUCUGCAAUCGGUCAGUGCAGU .....(((((((....((((.(....).))))((((((.....(((((((((....)-))))(((((((((......))).))))))........)))).)))))).......))))))) ( -25.80) >consensus UUUCCGCUGCACCUUAUAGAAAAAGUCAUCCAUUGUAGCCAUUGUUCUAUUUAUACAAAAUAUAUAUUCCUAUUUGGUGGUGAUAUGACGUGUGUGAACUCUGCAAUCGGUCGGUGCAGU .....((((((((.....((.....))..(((((((((.....((((....(((((...((((.....((.....)).....))))...))))).)))).))))))).))..)))))))) (-24.64 = -25.52 + 0.88)

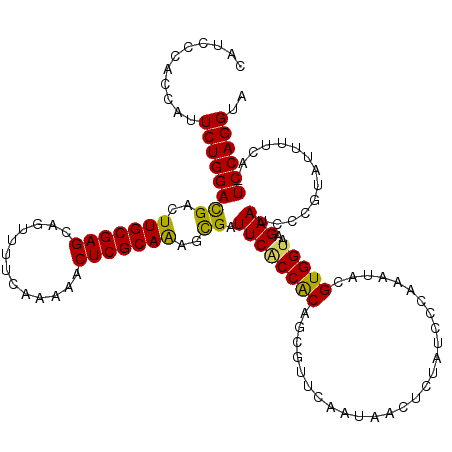
| Location | 20,482,740 – 20,482,859 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -21.67 |
| Energy contribution | -20.91 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20482740 119 + 22407834 CAUCCCACCAUUCUGGACGACUUGCGAGCAGUUUUCAAAAACUCGCAAAGUGAUUCUCCACAGCGUUCAAUAACUCUAUCCCAAAUACGUGGUAAGAAUUCCGUUUUUUCA-UCCAGGUA ...........((((((.((.(((((((.............))))))).(..(((((..((.((((....................)))).)).)))))..)......)).-)))))).. ( -24.47) >DroSec_CAF1 35663 119 + 1 CAUCCCAACAUUCUGGACGACUUGCGAGCAGUUUUCAAAAACUCGCAAAGCGAUUCGCCGCAUCGUUCAAUAACUCUAUCCCAAAUACGUGGUAAGAAUCCGCUCUUUUCA-UCCAGGUA ...........((((((.((.(((((((.............)))))))(((((((((((((...........................)))))..)))).))))....)).-)))))).. ( -27.75) >DroSim_CAF1 39486 119 + 1 CAUCCCACCAUUCUGGACGACUUGCGAGCAGUUUUCAAAAACUCGCAAAGCGAUUCGCCGCAUCGUUCAAUAACUCUAUCCCAAAUACGUGGUAAGAAUCCGCUCUUUUCA-UCCAGGUA ...........((((((.((.(((((((.............)))))))(((((((((((((...........................)))))..)))).))))....)).-)))))).. ( -27.75) >DroEre_CAF1 38061 120 + 1 CAUCCGACCAUUCUGGACGACUUGCGAGAAGUUUUCAAAAACUCGCAGAGCGAUUCACCACAGCGUUCCAUAACGCUAUCCCAAAUACGUGGUAAGAACCCCGAACUGUUGUUCCAGGUA ..((..(((((..(((.((..(((((((.............)))))))..)).........((((((....))))))...))).....)))))..))(((..((((....))))..))). ( -31.32) >DroYak_CAF1 35413 119 + 1 CAUCCCACCAUUCUGGAUGACUUGCGAGCAGUUUUUAAAAACUCGCAGAGCGAUUCACCACAGCGUUCAAUAACUCUGUCCCAAAUACGUGGUAGGAACAUCGAAUUUUUA-UCCAGGUA ...........(((((((((.(((((((.............)))))))..((((...((((((.(((....))).)))).(((......)))..))...)))).....)))-)))))).. ( -28.52) >consensus CAUCCCACCAUUCUGGACGACUUGCGAGCAGUUUUCAAAAACUCGCAAAGCGAUUCACCACAGCGUUCAAUAACUCUAUCCCAAAUACGUGGUAAGAAUCCCGUAUUUUCA_UCCAGGUA ...........((((((((..(((((((.............)))))))..)).((((((((...........................)))))..)))..............)))))).. (-21.67 = -20.91 + -0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:11 2006