
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,481,251 – 20,481,370 |
| Length | 119 |
| Max. P | 0.865723 |

| Location | 20,481,251 – 20,481,370 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.45 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -26.64 |
| Energy contribution | -26.80 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
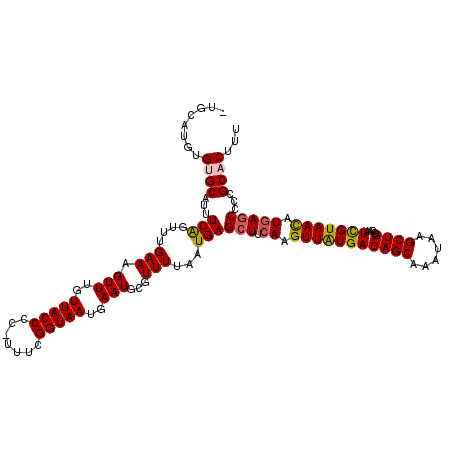
>2L_DroMel_CAF1 20481251 119 + 22407834 GUGCAUGUGUGCAUUUCAGUUUGAAAGUUUGUUACCCC-UUUCGGUAAUGAAUGCGUUUUAAUGAGCUUCAAGUUAUGACAGCAAAUAAGCUGGCAAUCGUAACAUGAGCCUCGCACUUU ((((......((.((((.....))))(((..(((((..-....)))))..)))))........(((.((((.(((((((((((......))))....))))))).)))).)))))))... ( -36.30) >DroSec_CAF1 34144 115 + 1 ----AUGUGUGCGUUUCAGUUUGAAAGUUUGUUACCCC-UUUCGGUAAUGAAUGCGUUUUAAUGAGCUUCAAGUUAUGACAGCAAAUAAGCUGGCAAUCGUAACAUGAGCCCCGCACUUU ----....(((((..(((....(((.(((..(((((..-....)))))..)))...)))...)))(((.((.(((((((((((......))))....))))))).)))))..)))))... ( -33.90) >DroSim_CAF1 37965 115 + 1 ----AUGUGUGCGUUUCAGUUUGAAAGUUUGUUACCCC-UUUCGGUAAUGAAUGCGUUUUAAUGAGCUUCAAGUUAUGACAGCAAAUAAGCUGGCAAUCGUAACAUGAGCCCCGCACUUU ----....(((((..(((....(((.(((..(((((..-....)))))..)))...)))...)))(((.((.(((((((((((......))))....))))))).)))))..)))))... ( -33.90) >DroEre_CAF1 36227 119 + 1 GUGCAUGUGUGCAGUUCGGUUUGAAAGUUUGUUACCCC-UUUCGGUAAUGAAUGCGUUUUAAUGAGGUUCAAGUUGUGACAGCAAAUAAGCUGGCAAUCGUAACAUAAGCCCCGCUCUUU (..(....)..)(((..((((((...(((..(((((..-....)))))..))).............(((...(((((..((((......)))))))))...))).))))))..))).... ( -26.60) >DroYak_CAF1 33734 120 + 1 AUGCAUGUGUGCAUUUCAGUUUGAAAGUUUGUUACCCCGCUUCGGUAAUGAAUGCGUUUUAAUGAGCUUCAAGUUAUGACAGCAAAUAAGCUGGCAAUUGUAAUAUGAGCCCCGCUCUUU .((((....(((...(((.((((((.(((..(((...(((((((....)))).)))...)))..)))))))))...)))((((......)))))))..))))....((((...))))... ( -31.20) >consensus _UGCAUGUGUGCAUUUCAGUUUGAAAGUUUGUUACCCC_UUUCGGUAAUGAAUGCGUUUUAAUGAGCUUCAAGUUAUGACAGCAAAUAAGCUGGCAAUCGUAACAUGAGCCCCGCACUUU ........((((...(((....(((.(((..(((((.......)))))..)))...)))...)))(((.((.(((((((((((......))))....))))))).)))))...))))... (-26.64 = -26.80 + 0.16)

| Location | 20,481,251 – 20,481,370 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.45 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
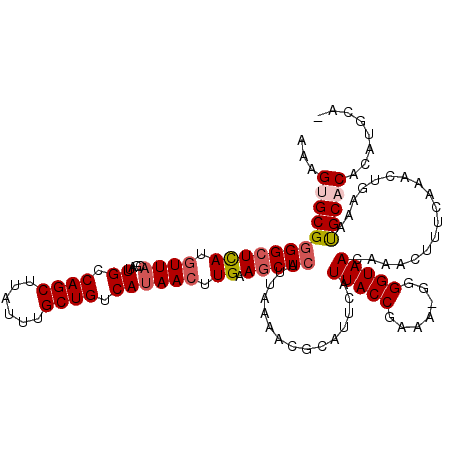
>2L_DroMel_CAF1 20481251 119 - 22407834 AAAGUGCGAGGCUCAUGUUACGAUUGCCAGCUUAUUUGCUGUCAUAACUUGAAGCUCAUUAAAACGCAUUCAUUACCGAAA-GGGGUAACAAACUUUCAAACUGAAAUGCACACAUGCAC ...(((((.......((((((((.(((((((......))))(((.....))).............))).))....((....-)).))))))...((((.....)))))))))........ ( -29.50) >DroSec_CAF1 34144 115 - 1 AAAGUGCGGGGCUCAUGUUACGAUUGCCAGCUUAUUUGCUGUCAUAACUUGAAGCUCAUUAAAACGCAUUCAUUACCGAAA-GGGGUAACAAACUUUCAAACUGAAACGCACACAU---- ...(((((.......((((((((.(((((((......))))(((.....))).............))).))....((....-)).))))))...((((.....)))))))))....---- ( -31.60) >DroSim_CAF1 37965 115 - 1 AAAGUGCGGGGCUCAUGUUACGAUUGCCAGCUUAUUUGCUGUCAUAACUUGAAGCUCAUUAAAACGCAUUCAUUACCGAAA-GGGGUAACAAACUUUCAAACUGAAACGCACACAU---- ...(((((.......((((((((.(((((((......))))(((.....))).............))).))....((....-)).))))))...((((.....)))))))))....---- ( -31.60) >DroEre_CAF1 36227 119 - 1 AAAGAGCGGGGCUUAUGUUACGAUUGCCAGCUUAUUUGCUGUCACAACUUGAACCUCAUUAAAACGCAUUCAUUACCGAAA-GGGGUAACAAACUUUCAAACCGAACUGCACACAUGCAC .....((((......((((((((.(((((((......))))(((.....))).............))).))....((....-)).))))))....(((.....))))))).......... ( -26.40) >DroYak_CAF1 33734 120 - 1 AAAGAGCGGGGCUCAUAUUACAAUUGCCAGCUUAUUUGCUGUCAUAACUUGAAGCUCAUUAAAACGCAUUCAUUACCGAAGCGGGGUAACAAACUUUCAAACUGAAAUGCACACAUGCAU .....((((((((((..(((....((.((((......)))).)))))..)).)))))........(((((..(((((.......))))).......((.....))))))).....))).. ( -23.90) >consensus AAAGUGCGGGGCUCAUGUUACGAUUGCCAGCUUAUUUGCUGUCAUAACUUGAAGCUCAUUAAAACGCAUUCAUUACCGAAA_GGGGUAACAAACUUUCAAACUGAAAUGCACACAUGCA_ ...((((((((((((.((((....((.((((......)))).)))))).)).)))))...............(((((.......)))))..................)))))........ (-22.02 = -22.82 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:08 2006