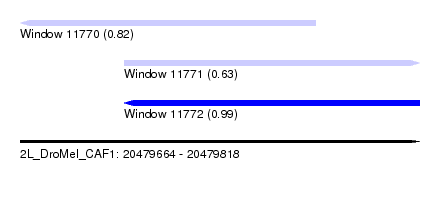
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,479,664 – 20,479,818 |
| Length | 154 |
| Max. P | 0.988824 |

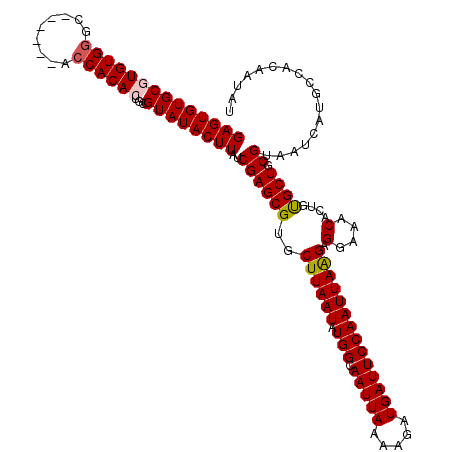
| Location | 20,479,664 – 20,479,778 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.78 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20479664 114 - 22407834 GAGUGUGCGUGUGGGC------ACCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAAUUAAGAGGAAACACUGUGCUGCGUAAUCAUGCCACAAUAU ((((((((((((((..------..).))).))))))))))...(.(((((.(((...(((((......((.((.((((........)))).)))).)))))..))).))))))....... ( -31.00) >DroSec_CAF1 32599 114 - 1 GAGUGUGCGUGUGAGC------ACCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAAUUAAGAGGAAACACUGUGCUGCGUAAUCAUGCCACAAUAU ..(((.(((((...((------(.((((.((.......(((((.....((((......)))).......)))))((((........)))).)).)))).))).....))))))))..... ( -29.90) >DroSim_CAF1 36418 114 - 1 GAGUGUGCGUGUGGGC------ACCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAAUUAAGAGGAAACACUGUGCUGCGUAAUCAUGCCACAAUAU ((((((((((((((..------..).))).))))))))))...(.(((((.(((...(((((......((.((.((((........)))).)))).)))))..))).))))))....... ( -31.00) >DroEre_CAF1 34650 112 - 1 GAGUGUGCAUGUGGG--------CCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAAUUAGGAGGAAACACUGCGCUGCGUAAUCACGGCACAAUAU ...(((((.(((((.--------..((.(.(((((...(((((.....((((......)))).......))))).(((.....)))(....)..))))).).))..)))))))))).... ( -30.50) >DroYak_CAF1 32116 120 - 1 GAGUGUGCAUGUGGGCACCGGCACCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAAUUAAGAGGAAACACUGUGCUGCGUAAUCAUGGCACAAUAU .(((((((((((((((....)).))))).)))......(((((.....((((......)))).......)))))((((........)))))))))(((((..(....)..)))))..... ( -34.10) >consensus GAGUGUGCGUGUGGGC______ACCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAAUUAAGAGGAAACACUGUGCUGCGUAAUCAUGCCACAAUAU ((((((((((((((.........))))))...))))))))..((((((..((((((.(((.(((((.....)))))))))))))).(....)....)))).))................. (-27.50 = -27.78 + 0.28)

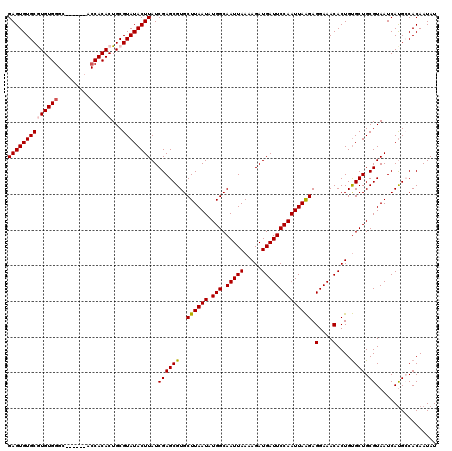
| Location | 20,479,704 – 20,479,818 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.02 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.10 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20479704 114 + 22407834 UUGGAAUCAUCUUUUAAUUGCCAUAUUAAGCACGCUCGAUAAGUAUACGCAGUGUGGU------GCCCACACGCACACUCCUCGCUCGCAUCCUGCACACACACUAGCACAAUGGCAAGC .................(((((((((..((....))..))).......((((((((((------((......))))...........((.....))...)))))).))....)))))).. ( -28.70) >DroSec_CAF1 32639 114 + 1 UUGGAAUCAUCUUUUAAUUGCCAUAUUAAGCACGCUCGAUAAGUAUACGCAGUGUGGU------GCUCACACGCACACUCCUCGCUCGCAUCCUGCACACACACUAGCACAAUGGCAAGC .................(((((((((..((....))..))).......((((((((((------((......))))...........((.....))...)))))).))....)))))).. ( -29.20) >DroSim_CAF1 36458 114 + 1 UUGGAAUCAUCUUUUAAUUGCCAUAUUAAGCACGCUCGAUAAGUAUACGCAGUGUGGU------GCCCACACGCACACUCCUCGCUCGCAUCCUGCACACACACUAGCACAAUGGCAAGC .................(((((((((..((....))..))).......((((((((((------((......))))...........((.....))...)))))).))....)))))).. ( -28.70) >DroEre_CAF1 34690 112 + 1 UUGGAAUCAUCUUUUAAUUGCCAUAUUAAGCACGCUCGAUAAGUAUACGCAGUGUGG--------CCCACAUGCACACUCUUCGCUCGCAUCCUGCACACACACAAGCACAAUGGCACGC ..................((((((((..((....))..))).......((.((((((--------(....((((.............))))...)).)))))....))....)))))... ( -20.82) >DroYak_CAF1 32156 120 + 1 UUGGAAUCAUCUUUUAAUUGCCAUAUUAAGCACGCUCGAUAAGUAUACGCAGUGUGGUGCCGGUGCCCACAUGCACACUCUUCGCUCGCAUCCUGCACACACACAAGCACAAUGGCAAGC .................(((((((((..((....))..))).......((.(((((((((.(((((......((.........))..))).)).)))).)))))..))....)))))).. ( -31.10) >consensus UUGGAAUCAUCUUUUAAUUGCCAUAUUAAGCACGCUCGAUAAGUAUACGCAGUGUGGU______GCCCACACGCACACUCCUCGCUCGCAUCCUGCACACACACUAGCACAAUGGCAAGC .................(((((((.....((..(..(((..(((....((.((((((.........))))))))..)))..)))..)((.....))..........))...))))))).. (-23.30 = -23.10 + -0.20)

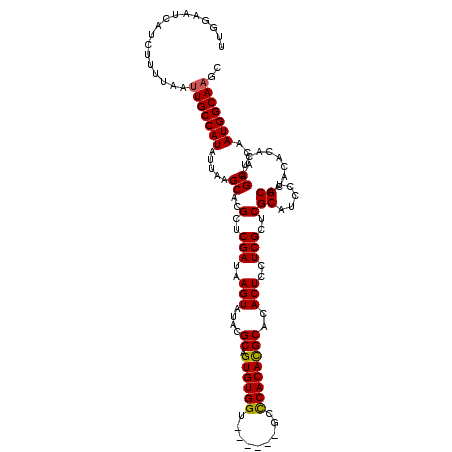

| Location | 20,479,704 – 20,479,818 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.02 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -35.92 |
| Energy contribution | -36.72 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20479704 114 - 22407834 GCUUGCCAUUGUGCUAGUGUGUGUGCAGGAUGCGAGCGAGGAGUGUGCGUGUGGGC------ACCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAA ((((((.(((.(((..........))).)))))))))..(((((((((((((((..------..).))).))))))))(((((.....((((......)))).......))))).))).. ( -38.80) >DroSec_CAF1 32639 114 - 1 GCUUGCCAUUGUGCUAGUGUGUGUGCAGGAUGCGAGCGAGGAGUGUGCGUGUGAGC------ACCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAA ((((((.(((.(((..........))).)))))))))..((((((((((((((...------....))).))))))))(((((.....((((......)))).......))))).))).. ( -37.20) >DroSim_CAF1 36458 114 - 1 GCUUGCCAUUGUGCUAGUGUGUGUGCAGGAUGCGAGCGAGGAGUGUGCGUGUGGGC------ACCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAA ((((((.(((.(((..........))).)))))))))..(((((((((((((((..------..).))).))))))))(((((.....((((......)))).......))))).))).. ( -38.80) >DroEre_CAF1 34690 112 - 1 GCGUGCCAUUGUGCUUGUGUGUGUGCAGGAUGCGAGCGAAGAGUGUGCAUGUGGG--------CCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAA (((..((((.......))).)..))).(((.(((..(((.((((((((..(((..--------...)))...)))))))).)))..)))(((......)))..............))).. ( -34.50) >DroYak_CAF1 32156 120 - 1 GCUUGCCAUUGUGCUUGUGUGUGUGCAGGAUGCGAGCGAAGAGUGUGCAUGUGGGCACCGGCACCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAA ..(((((((.(((((((.((((((((((.(((((.((.....)).)))))((((((....)).)))).))))))))))....)))))..)).....)))))))................. ( -39.80) >consensus GCUUGCCAUUGUGCUAGUGUGUGUGCAGGAUGCGAGCGAGGAGUGUGCGUGUGGGC______ACCACACUGCGUAUACUUAUCGAGCGUGCUUAAUAUGGCAAUUAAAAGAUGAUUCCAA ..(((((((((..(........)..))(.((((...(((.((((((((((((((.........))))))...)))))))).))).)))).).....)))))))................. (-35.92 = -36.72 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:34:04 2006