
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,477,396 – 20,477,594 |
| Length | 198 |
| Max. P | 0.999516 |

| Location | 20,477,396 – 20,477,516 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -47.32 |
| Consensus MFE | -38.72 |
| Energy contribution | -39.80 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20477396 120 + 22407834 CGGGCGGGGGUCCCUGGGAACGGCGGCCACACAGCAGUGUACUCUGUGUACUUGGCUCAUGCAUAACGCAAAUGCUGGAAACGCCGCAAGGAGCCGCAUCGGCAUCCUUGCCAGUUUCGU ....((((....))))(((((((((.((((((((.((....))))))))....(((...(((.....)))...)))))...))))(((((((((((...)))).)))))))..))))).. ( -52.00) >DroSec_CAF1 30403 120 + 1 CGGGCGGGGGUCCCUGGCAACGGCGGCCACACAGCAGUGUACUCUGUGUACUUUGCUCAUGCAUAACGCAAAUGCUGGAAACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGU .(((((((((((.(((....))).))))((((((.((....))))))))..)))))))..((((.......))))..(((((...((((((((((.....))).)))))))..))))).. ( -51.10) >DroSim_CAF1 34180 120 + 1 CGGGCGGGGGUCCCUGGAAGCGGCGGCCACACAGCAGUGUACUCUGUGUACUUGGCUCAUGCAUAACGCAAAUGCUGGAAACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGU ....((((....))))(((((((((.((((((((.((....))))))))....(((...(((.....)))...)))))...))))((((((((((.....))).)))))))..))))).. ( -51.70) >DroEre_CAF1 32497 114 + 1 CGGGCGGGGGUCCAUUGAAAUGGCGGCCACGCAGCA-AGUACAC-----ACUUGGCCCAUUCAUAACGCAAAUGCUGGAGACGCCGCAAGGAGCCGCAGCGGCAUCCUUGCCAGUUUCGU .((((....))))...(((((((((.(((.(((.((-(((....-----)))))((...........))...))))))...))))(((((((((((...)))).)))))))..))))).. ( -43.90) >DroYak_CAF1 29594 115 + 1 CGGGCGGGGGUCCAUUGAAAUAGCGGCCACACAACAGAGUACAC-----ACUUGGCUCAUGCAUAACGCAAAUGCUGGAAACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGU .((((....))))...(((((.(((.(((((.....((((.((.-----...)))))).(((.....)))..)).)))...))).((((((((((.....))).)))))))..))))).. ( -37.90) >consensus CGGGCGGGGGUCCCUGGAAACGGCGGCCACACAGCAGUGUACUCUGUGUACUUGGCUCAUGCAUAACGCAAAUGCUGGAAACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGU .((((....))))...(((((((((......(((((..((((.....))))........(((.....)))..)))))....))))((((((((((.....))).)))))))..))))).. (-38.72 = -39.80 + 1.08)

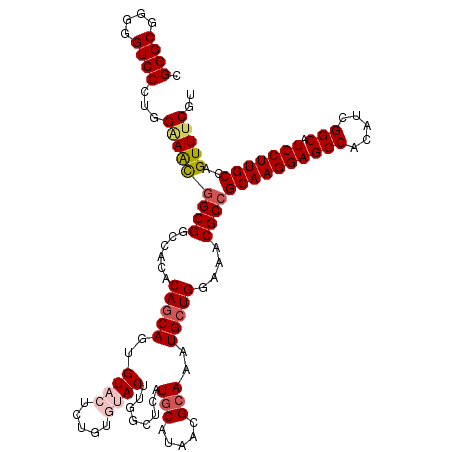
| Location | 20,477,436 – 20,477,554 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -36.98 |
| Energy contribution | -36.86 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20477436 118 + 22407834 ACUCUGUGUACUUGGCUCAUGCAUAACGCAAAUGCUGGAAACGCCGCAAGGAGCCGCAUCGGCAUCCUUGCCAGUUUCGUCCUCGUCGAGGUCCCAGCAUGUCCAC--GCAAUAACGAUU ....(((((....(((...(((.....))).(((((((((((...(((((((((((...)))).)))))))..)))))(.((((...)))).).))))))))).))--)))......... ( -42.20) >DroSec_CAF1 30443 120 + 1 ACUCUGUGUACUUUGCUCAUGCAUAACGCAAAUGCUGGAAACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGUCCUCGUGGAGGUCCCAGCAUGUCCAUGUGCAAUAACGAUU ......(((...((((.(((((.....))..(((((((((((...((((((((((.....))).)))))))..)))))(.((((...)))).).))))))....))).))))..)))... ( -42.10) >DroSim_CAF1 34220 118 + 1 ACUCUGUGUACUUGGCUCAUGCAUAACGCAAAUGCUGGAAACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGUCCUCGUGGAGGUCCCAGCAUGUCCAC--GCAAUAACGAUU ....(((((....(((...(((.....))).(((((((((((...((((((((((.....))).)))))))..)))))(.((((...)))).).))))))))).))--)))......... ( -41.40) >DroEre_CAF1 32536 112 + 1 ACAC-----ACUUGGCCCAUUCAUAACGCAAAUGCUGGAGACGCCGCAAGGAGCCGCAGCGGCAUCCUUGCCAGUUUCGUCCUC-CGGAGGUCCCAGCAUGACCAC--GCAAUAACGAUU ....-----..................((..(((((((.(((...(((((((((((...)))).)))))))...(((((.....-)))))))))))))))......--)).......... ( -35.50) >DroYak_CAF1 29634 113 + 1 ACAC-----ACUUGGCUCAUGCAUAACGCAAAUGCUGGAAACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGUCCUCGUGGAGGUCCCAGCGUGUCCAC--GCAAUAACGAUU ....-----.....((...(((.....))).(((((((((((...((((((((((.....))).)))))))..)))))(.((((...)))).).))))))......--)).......... ( -37.80) >consensus ACUCUGUGUACUUGGCUCAUGCAUAACGCAAAUGCUGGAAACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGUCCUCGUGGAGGUCCCAGCAUGUCCAC__GCAAUAACGAUU ..............((...(((.....))).(((((((((((...((((((((((.....))).)))))))..)))))(.((((...)))).).))))))........)).......... (-36.98 = -36.86 + -0.12)

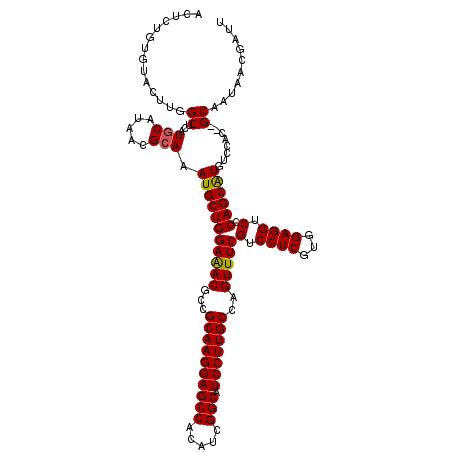
| Location | 20,477,436 – 20,477,554 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -41.94 |
| Consensus MFE | -36.18 |
| Energy contribution | -35.98 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20477436 118 - 22407834 AAUCGUUAUUGC--GUGGACAUGCUGGGACCUCGACGAGGACGAAACUGGCAAGGAUGCCGAUGCGGCUCCUUGCGGCGUUUCCAGCAUUUGCGUUAUGCAUGAGCCAAGUACACAGAGU ..(((((((.((--(..((..(((((...((((...))))..((((((.(((((((.((((...))))))))))).).))))))))))))..))).))).))))................ ( -43.10) >DroSec_CAF1 30443 120 - 1 AAUCGUUAUUGCACAUGGACAUGCUGGGACCUCCACGAGGACGAAACUGGCAAGGAUGCCGAUGUGGCUCCUUGCGGCGUUUCCAGCAUUUGCGUUAUGCAUGAGCAAAGUACACAGAGU ....(((..((((.(((.(.((((((...((((...))))..((((((.(((((((.(((.....)))))))))).).))))))))))).).)))..))))..))).............. ( -41.40) >DroSim_CAF1 34220 118 - 1 AAUCGUUAUUGC--GUGGACAUGCUGGGACCUCCACGAGGACGAAACUGGCAAGGAUGCCGAUGUGGCUCCUUGCGGCGUUUCCAGCAUUUGCGUUAUGCAUGAGCCAAGUACACAGAGU ..(((((((.((--(..((..(((((...((((...))))..((((((.(((((((.(((.....)))))))))).).))))))))))))..))).))).))))................ ( -41.90) >DroEre_CAF1 32536 112 - 1 AAUCGUUAUUGC--GUGGUCAUGCUGGGACCUCCG-GAGGACGAAACUGGCAAGGAUGCCGCUGCGGCUCCUUGCGGCGUCUCCAGCAUUUGCGUUAUGAAUGGGCCAAGU-----GUGU ..(((((...((--(..(..((((((((((..((.-.((.......)).(((((((.((((...))))))))))))).))).))))))))..)))....))))).......-----.... ( -42.00) >DroYak_CAF1 29634 113 - 1 AAUCGUUAUUGC--GUGGACACGCUGGGACCUCCACGAGGACGAAACUGGCAAGGAUGCCGAUGUGGCUCCUUGCGGCGUUUCCAGCAUUUGCGUUAUGCAUGAGCCAAGU-----GUGU ...(((...(((--.((((.(((((....((((...)))).........(((((((.(((.....))))))))))))))).)))))))...)))..((((((.......))-----)))) ( -41.30) >consensus AAUCGUUAUUGC__GUGGACAUGCUGGGACCUCCACGAGGACGAAACUGGCAAGGAUGCCGAUGUGGCUCCUUGCGGCGUUUCCAGCAUUUGCGUUAUGCAUGAGCCAAGUACACAGAGU ...(((...(((...((((.(((((....((((...)))).........(((((((.((((...)))))))))))))))).)))))))...))).......................... (-36.18 = -35.98 + -0.20)


| Location | 20,477,476 – 20,477,594 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.79 |
| Mean single sequence MFE | -37.85 |
| Consensus MFE | -35.04 |
| Energy contribution | -35.08 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20477476 118 + 22407834 ACGCCGCAAGGAGCCGCAUCGGCAUCCUUGCCAGUUUCGUCCUCGUCGAGGUCCCAGCAUGUCCAC--GCAAUAACGAUUGCGGGCAUAAUGCCCAUAAUGUCCUCAUAAUUACUGCCCA .....(((((((((((...)))).)))))))((((..((....))..((((.....((((......--(((((....)))))((((.....))))...))))))))......)))).... ( -37.90) >DroSec_CAF1 30483 120 + 1 ACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGUCCUCGUGGAGGUCCCAGCAUGUCCAUGUGCAAUAACGAUUGCGGGCAUAAUGCCCAUAAUGUCCUCAUAAUUACUGCCCA .....((((((((((.....))).)))))))((((........((((((.((......)).)))))).(((((....)))))((((.....)))).................)))).... ( -38.30) >DroSim_CAF1 34260 118 + 1 ACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGUCCUCGUGGAGGUCCCAGCAUGUCCAC--GCAAUAACGAUUGCGGGCAUAAUGCCCAUAAUGUCCUCGUAAUUACUGCCCA .....((((((((((.....))).)))))))((((..((....((((((.((......)).)))))--(((((....)))))((((.....)))).....)....)).....)))).... ( -39.70) >DroEre_CAF1 32571 117 + 1 ACGCCGCAAGGAGCCGCAGCGGCAUCCUUGCCAGUUUCGUCCUC-CGGAGGUCCCAGCAUGACCAC--GCAAUAACGAUUGCGGGCAUAAUGCCCAUAAUGUCCUCAUAAUUACUGCCCA .....(((((((((((...)))).)))))))((((.........-.(((((((.......))))..--(((((....)))))((((.....))))......)))........)))).... ( -38.57) >DroYak_CAF1 29669 118 + 1 ACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGUCCUCGUGGAGGUCCCAGCGUGUCCAC--GCAAUAACGAUUGCGGCCAUAAUGCCCAUAAUGUCCUCAUAAUUACUGCCCA ..(((((((((..((.....((((....)))).......(((....)))))..)).((((....))--))........)))))))................................... ( -34.80) >consensus ACGCCGCAAGGAGCCACAUCGGCAUCCUUGCCAGUUUCGUCCUCGUGGAGGUCCCAGCAUGUCCAC__GCAAUAACGAUUGCGGGCAUAAUGCCCAUAAUGUCCUCAUAAUUACUGCCCA .....((((((((((.....))).)))))))((((...(.((((...)))).)...((((........(((((....)))))((((.....))))...))))..........)))).... (-35.04 = -35.08 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:58 2006