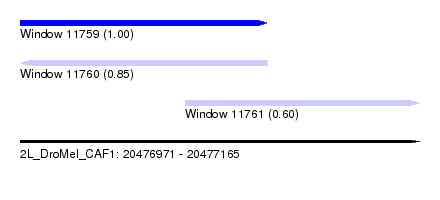
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,476,971 – 20,477,165 |
| Length | 194 |
| Max. P | 0.995660 |

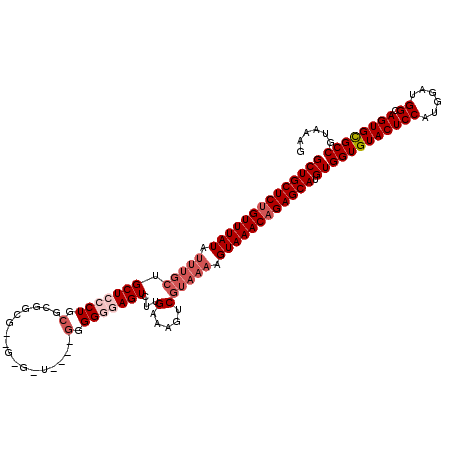
| Location | 20,476,971 – 20,477,091 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.41 |
| Mean single sequence MFE | -44.74 |
| Consensus MFE | -34.17 |
| Energy contribution | -36.13 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20476971 120 + 22407834 GCUGCUCUGUUUAUAUUUGCUGCUCCCUGCGCGGCGCGGCGGUUGGUGGGGGGAGUCUUGAAAGUCGUAAAAGUAAACAGAGCAUGUGGUGUACUCCAUGGAUGGCAGUGCGCCGUAAAG ((((((((((((((.(((((..(((((..(.((((......)))))..))))).(.((....)).)))))).)))))))))))).))(((((((((((....))).))))))))...... ( -53.20) >DroSec_CAF1 29981 116 + 1 GCUGCUCUGUUUAUAUUUGCUGCUCCCUGCGCGUUGAAGGGGU----UGGGGGAGUCUUGAAAAACGUAAAAGUAAACAGAGCAUGUGGUGUACUCCAUGGAUGGCAGUGCGCCGUAAAG ((((((((((((((.(((((.(((((((((.(.......).))----..))))))).((....)).))))).)))))))))))).))(((((((((((....))).))))))))...... ( -47.70) >DroSim_CAF1 33758 116 + 1 GCUGCUCUGUUUAUAUUUGCUGCUCCCUGCGCGUUGAAGGGUU----GGGGGGAGUCUUGAAAGUCGUAAAAGUAAACAGAGCAUGUGGUGUACUCCAUGGAUGGCAGUGCGCCGUAAAG ((((((((((((((.(((((..(((((.((.(......).)).----)))))..(.((....)).)))))).)))))))))))).))(((((((((((....))).))))))))...... ( -48.80) >DroEre_CAF1 32101 99 + 1 GCUGCUCCGUUUAUG-----CGCUCCCUGCGCGGCG-----------G-GGAAAGUCUUGAAAGUCGUAAAAGUAAACAGAGCAUGUGGUGUACUCCAU----GGCAGUGUGCCGUAAAG (((((((.((((((.-----.(((((((((...)))-----------)-)))..(.((....)).)))....)))))).))))).))((..(((((...----.).))))..))...... ( -33.10) >DroYak_CAF1 29190 105 + 1 GCUGCUCUGUUUAUAUUUGCCGCUCCCUGCGCGGCG-----------GGGGAAAGUCUUGAAAGUCGUAAAAGUAAACAGAGCAUGUGGUGUACUCCAU----GGCAGUGUGCCGUAAAG ((((((((((((((.(((((.(((((((((...)))-----------)))............))).))))).)))))))))))).))((..(((((...----.).))))..))...... ( -40.90) >consensus GCUGCUCUGUUUAUAUUUGCUGCUCCCUGCGCGGCG__G_G_U____GGGGGGAGUCUUGAAAGUCGUAAAAGUAAACAGAGCAUGUGGUGUACUCCAUGGAUGGCAGUGCGCCGUAAAG ((((((((((((((.(((((.(((((((.(.................).)))))))...(.....)))))).)))))))))))).))((((((((((......)).))))))))...... (-34.17 = -36.13 + 1.96)

| Location | 20,476,971 – 20,477,091 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.41 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -23.84 |
| Energy contribution | -23.60 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20476971 120 - 22407834 CUUUACGGCGCACUGCCAUCCAUGGAGUACACCACAUGCUCUGUUUACUUUUACGACUUUCAAGACUCCCCCCACCAACCGCCGCGCCGCGCAGGGAGCAGCAAAUAUAAACAGAGCAGC ......((.(.(((.(((....)))))).).))...(((((((((((..(((.((.....)..(.(((((.........(((......)))..)))))).).)))..))))))))))).. ( -31.10) >DroSec_CAF1 29981 116 - 1 CUUUACGGCGCACUGCCAUCCAUGGAGUACACCACAUGCUCUGUUUACUUUUACGUUUUUCAAGACUCCCCCA----ACCCCUUCAACGCGCAGGGAGCAGCAAAUAUAAACAGAGCAGC ......((.(.(((.(((....)))))).).))...(((((((((((..(((.(((((....)))).......----..((((.(.....).))))....).)))..))))))))))).. ( -29.50) >DroSim_CAF1 33758 116 - 1 CUUUACGGCGCACUGCCAUCCAUGGAGUACACCACAUGCUCUGUUUACUUUUACGACUUUCAAGACUCCCCCC----AACCCUUCAACGCGCAGGGAGCAGCAAAUAUAAACAGAGCAGC ......((.(.(((.(((....)))))).).))...(((((((((((..(((.((.....)..(.(((((...----................)))))).).)))..))))))))))).. ( -29.21) >DroEre_CAF1 32101 99 - 1 CUUUACGGCACACUGCC----AUGGAGUACACCACAUGCUCUGUUUACUUUUACGACUUUCAAGACUUUCC-C-----------CGCCGCGCAGGGAGCG-----CAUAAACGGAGCAGC (((((.(((.....)))----.))))).........(((((((((((.......(.....)..(.(.((((-(-----------.((...)).))))).)-----).))))))))))).. ( -28.20) >DroYak_CAF1 29190 105 - 1 CUUUACGGCACACUGCC----AUGGAGUACACCACAUGCUCUGUUUACUUUUACGACUUUCAAGACUUUCCCC-----------CGCCGCGCAGGGAGCGGCAAAUAUAAACAGAGCAGC (((((.(((.....)))----.))))).........(((((((((((..(((..(.....)............-----------.(((((.......))))))))..))))))))))).. ( -33.00) >consensus CUUUACGGCGCACUGCCAUCCAUGGAGUACACCACAUGCUCUGUUUACUUUUACGACUUUCAAGACUCCCCCC____A_C_C__CGCCGCGCAGGGAGCAGCAAAUAUAAACAGAGCAGC ......((.(.(((.((......))))).).))...(((((((((((.......(.....)..(.(((((.......................))))))........))))))))))).. (-23.84 = -23.60 + -0.24)

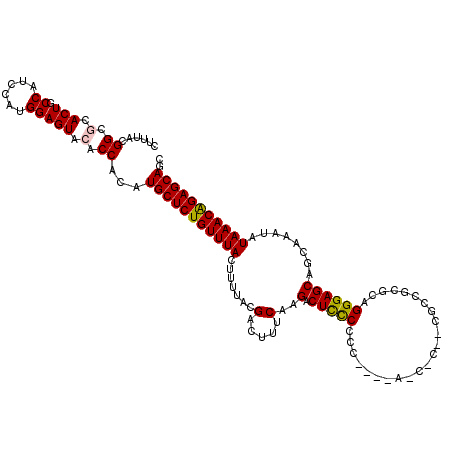
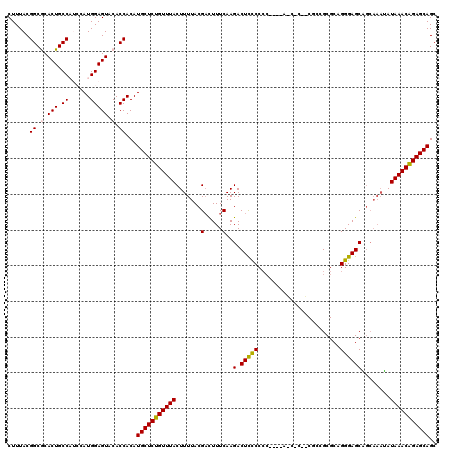
| Location | 20,477,051 – 20,477,165 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.06 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.12 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20477051 114 + 22407834 AGCAUGUGGUGUACUCCAUGGAUGGCAGUGCGCCGUAAAGCGCUUAAUUUUUUGU-----UGCCCUUCGCUGGCUCCAAUUUGAAGGAAGGUGAGAAAUAUGGCGCUCUGGCUGCCC-UG .(((...(((((((((((....))).))))))))((..((((((..((((((..(-----(..((((((.((....))...))))))..))..))))))..))))))...)))))..-.. ( -41.80) >DroSec_CAF1 30057 113 + 1 AGCAUGUGGUGUACUCCAUGGAUGGCAGUGCGCCGUAAAGCGCUUAAUUUUUUGU-----UGCCCUUCGCCGGCUCCAAUUUGG-GGAAGGUGAGAAAUAUGGCGCUCUGGCUGCCC-UG ..((((..(....)..))))...((((((((((((((..(((..(((....))).-----)))..((((((..((((.....))-))..))))))...))))))))....)))))).-.. ( -42.00) >DroSim_CAF1 33834 113 + 1 AGCAUGUGGUGUACUCCAUGGAUGGCAGUGCGCCGUAAAGCGCUUAAUUUUUUGU-----UGCCCUUCGCCGGCUCCAAUUUGG-GGAAGGUGAGAAAUAUGGCGCUCUGGCUGCCC-UG ..((((..(....)..))))...((((((((((((((..(((..(((....))).-----)))..((((((..((((.....))-))..))))))...))))))))....)))))).-.. ( -42.00) >DroEre_CAF1 32164 110 + 1 AGCAUGUGGUGUACUCCAU----GGCAGUGUGCCGUAAAGCGCUUAAUUUUUUGU-----UGCCCUUCGCUGGCUCCAAUUUGG-GGAAGGUGAGAAAAAUGGCGCACUGGCUGCCACCG .(((.((((......))))----(.(((((((((((...(((..(((....))).-----)))..((((((..((((.....))-))..))))))....))))))))))).))))..... ( -39.70) >DroYak_CAF1 29259 109 + 1 AGCAUGUGGUGUACUCCAU----GGCAGUGUGCCGUAAAGCGCUUAAUUUUUUGUUGCCU------UCGCUGGCUCCAAUUUGG-GGAAGGUGAGAAAUAUGGCGCUCUGGCUGCCGCCG .((..((((......))))----((((((((((((((..(((..(((....))).))).(------(((((..((((.....))-))..))))))...))))))))....)))))))).. ( -40.00) >consensus AGCAUGUGGUGUACUCCAUGGAUGGCAGUGCGCCGUAAAGCGCUUAAUUUUUUGU_____UGCCCUUCGCUGGCUCCAAUUUGG_GGAAGGUGAGAAAUAUGGCGCUCUGGCUGCCC_UG .(((...((((((((((......)).))))))))((..((((((..(((((...............(((((..((((.....)).))..))))))))))..))))))...)))))..... (-31.00 = -31.12 + 0.12)

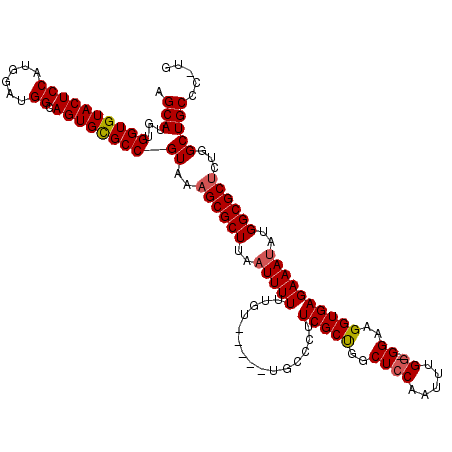
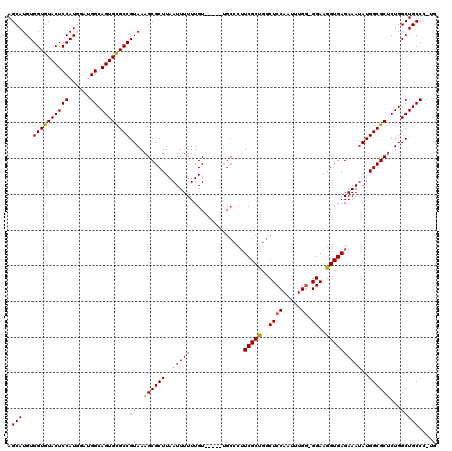
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:54 2006