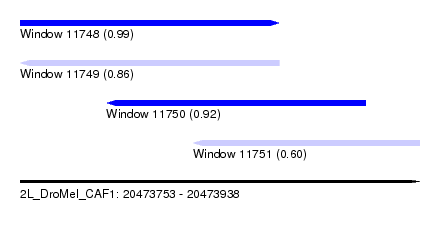
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,473,753 – 20,473,938 |
| Length | 185 |
| Max. P | 0.987170 |

| Location | 20,473,753 – 20,473,873 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -49.82 |
| Consensus MFE | -44.60 |
| Energy contribution | -44.08 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
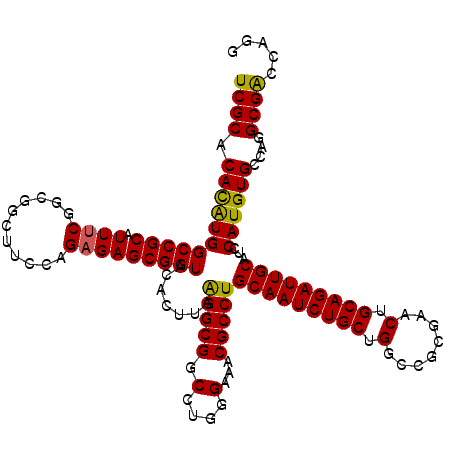
>2L_DroMel_CAF1 20473753 120 + 22407834 UCGCACACAUGGCCGCAUUUCGGCGACUUCCAGAGAGCGGUACACUUAAGGCGGCCUGGGAAACGCCUGCAAUCUGCUGGUCGCGAACUGCAGAUUGCAUCCAUGUGCCAGGCGACCAUG ((((.(((((((.........((((..((((((...((.((.........)).))))))))..))))((((((((((.(((.....))))))))))))).)))))))....))))..... ( -48.30) >DroSec_CAF1 26766 120 + 1 UCGCACACAUGGCCGCAUUUCGGCGGCUUCCAGAGAGCGGUACACUUAAGGCGGCCUGGGAAACGCCUGCAAUCUGCUGGCCGCGAACUGCAGAUUGCAUCCAUGUGCCAGGCGGCCAGG ((((.(.(..((((((......))))))....).).)))).........(((.((((((.(......((((((((((.(........).))))))))))......).)))))).)))... ( -52.40) >DroSim_CAF1 28733 120 + 1 UCGCACACAUGGCCGCAUUUCGGCGGCUUCCAGAGAGCGGUACACUUAAGGCGGCCUGGGAAACGCCUGCAAUCUGCUGGCCGCGAACUGCAGAUUGCAUCCAUGUGCCAGGCGGCCAGG ((((.(.(..((((((......))))))....).).)))).........(((.((((((.(......((((((((((.(........).))))))))))......).)))))).)))... ( -52.40) >DroEre_CAF1 29121 117 + 1 UCGCACAUGUGGCCGCAUUGCGUCCGCUUCCAGAGAGCGGUACACUUAGGGCGUCCUGGGAAACGCCUGCAAUCUGCUGGCCGCGAGCUGCAGAUUGCAUCCAUGUGCCAGGCGACC--- ..(((((((.(((........((((((((.....)))))).)).((((((....))))))....)))((((((((((.(((.....)))))))))))))..))))))).........--- ( -49.20) >DroYak_CAF1 26154 119 + 1 UCGCACACAUGGCCGCAUUUCGUCGGC-UCCAGAGAGCGGUACACUUAAGGCGUCCUGGGAAACGCCUGCAAUCUGCUGGCCGCGAACUGCAGAUUGCAUCCAUGUGCCAGGCGACCAAG ((((.(((((((((((.((((......-....))))))))).......((((((........))))))(((((((((.(........).)))))))))...))))))....))))..... ( -46.80) >consensus UCGCACACAUGGCCGCAUUUCGGCGGCUUCCAGAGAGCGGUACACUUAAGGCGGCCUGGGAAACGCCUGCAAUCUGCUGGCCGCGAACUGCAGAUUGCAUCCAUGUGCCAGGCGACCAGG ((((.(((((((((((.((((...........))))))))).......(((((.(....)...)))))(((((((((.(........).)))))))))...))))))....))))..... (-44.60 = -44.08 + -0.52)

| Location | 20,473,753 – 20,473,873 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -49.64 |
| Consensus MFE | -43.10 |
| Energy contribution | -43.82 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20473753 120 - 22407834 CAUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGACCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUACCGCUCUCUGGAAGUCGCCGAAAUGCGGCCAUGUGUGCGA .....((((...((((((((.((((((((((.(((...)))..)))))))))).(((((((...(((..((...((((...))))....))..)))...)))))))..)))))))))))) ( -47.10) >DroSec_CAF1 26766 120 - 1 CCUGGCCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUACCGCUCUCUGGAAGCCGCCGAAAUGCGGCCAUGUGUGCGA ...(((.((((((...(((..((((((((((.((.....))..))))))))))..)))...)))))).)))..........(((.(.(.....(((((......)))))...).).))). ( -49.40) >DroSim_CAF1 28733 120 - 1 CCUGGCCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUACCGCUCUCUGGAAGCCGCCGAAAUGCGGCCAUGUGUGCGA ...(((.((((((...(((..((((((((((.((.....))..))))))))))..)))...)))))).)))..........(((.(.(.....(((((......)))))...).).))). ( -49.40) >DroEre_CAF1 29121 117 - 1 ---GGUCGCCUGGCACAUGGAUGCAAUCUGCAGCUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGACGCCCUAAGUGUACCGCUCUCUGGAAGCGGACGCAAUGCGGCCACAUGUGCGA ---((((((...((((.(((.((((((((((.((.....))..))))))))))((((((......))))))))).)))).((((((....).))))).......)))))).......... ( -51.30) >DroYak_CAF1 26154 119 - 1 CUUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGACGCCUUAAGUGUACCGCUCUCUGGA-GCCGACGAAAUGCGGCCAUGUGUGCGA .....((((...((((((((.((((((((((.((.....))..)))))))))).(((((((...((((((.....)))).))((((....))-))....)))))))..)))))))))))) ( -51.00) >consensus CCUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUACCGCUCUCUGGAAGCCGCCGAAAUGCGGCCAUGUGUGCGA ......(((...((((((((.((((((((((.((.....))..)))))))))).(((((((...(((..((...((((...))))....))..)))...)))))))..))))))))))). (-43.10 = -43.82 + 0.72)



| Location | 20,473,793 – 20,473,913 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.32 |
| Mean single sequence MFE | -53.66 |
| Consensus MFE | -40.44 |
| Energy contribution | -42.90 |
| Covariance contribution | 2.46 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20473793 120 - 22407834 GGGCUGCGAGGCGUCGGGCGGGCACAUGGUCACAUGGUCGCAUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGACCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUA ((.(((.((((((((((((((.((..((..(....)..))..)).)))))))))....(..((((((((((.(((...)))..))))))))))..)))))).))).))............ ( -52.80) >DroSec_CAF1 26806 120 - 1 GGGAUGCGAGGCGUCGGGCGGGCAUAUGGUCGCAUGGUCGCCUGGCCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUA ...((((((((((((((((((.(((........))).))))))))).((((((...(((..((((((((((.((.....))..))))))))))..)))...)))))).)))))..)))). ( -56.40) >DroSim_CAF1 28773 120 - 1 GGGAUGCGAGGCGUCGGGCGGGCACAUGGUCGCCUGGUCGCCUGGCCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUA ...((((.(((((((((((((.(....).)))))))).)))))(((.((((((...(((..((((((((((.((.....))..))))))))))..)))...)))))).)))....)))). ( -61.10) >DroEre_CAF1 29161 93 - 1 ------AGAGG--A--CGU-GGCACAU----------------GGUCGCCUGGCACAUGGAUGCAAUCUGCAGCUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGACGCCCUAAGUGUA ------.....--.--...-.((((..----------------((.(((((((...(((..((((((((((.((.....))..))))))))))..)))...))))).)).))...)))). ( -38.20) >DroYak_CAF1 26193 119 - 1 UGGCUGCGAGGCGUCGGGC-GGCACAUGGUAACAUGGUCGCUUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGACGCCUUAAGUGUA .....((.(((((((((((-(((.((((....))))))))))))).))))).))((((...((((((((((.((.....))..))))))))))((((((......))))))....)))). ( -59.80) >consensus GGGAUGCGAGGCGUCGGGCGGGCACAUGGUCACAUGGUCGCCUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGGCCAGCAGAUUGCAGGCGUUUCCCAGGCCGCCUUAAGUGUA .......((((((..((.(((((.((((....))))...))))).))((((((...(((..((((((((((.((.....))..))))))))))..)))...))))))))))))....... (-40.44 = -42.90 + 2.46)



| Location | 20,473,833 – 20,473,938 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.55 |
| Mean single sequence MFE | -51.10 |
| Consensus MFE | -43.65 |
| Energy contribution | -44.90 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20473833 105 - 22407834 GUUGUGCCCUCGAUGGGCAGGAU---------------GUGGGCUGCGAGGCGUCGGGCGGGCACAUGGUCACAUGGUCGCAUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGA (((.(((((.....))))).)))---------------(((((((((...(.(((((((((.((..((..(....)..))..)).))))))))).)..((((...))))))))))))).. ( -49.10) >DroSec_CAF1 26846 120 - 1 GUUGUGCCCUCGAUGGGCAGGAUGUGAGGAUGUGCGAUGUGGGAUGCGAGGCGUCGGGCGGGCAUAUGGUCGCAUGGUCGCCUGGCCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGG (((.(((((.....))))).)))((((((((.((((.((((...(((.(((((((((((((.(((........))).)))))))).))))).)))))))..)))))))).....)))).. ( -52.30) >DroSim_CAF1 28813 120 - 1 GUUGUGCCCUCGAUGGGCAGGAUGUGAGGAUGCGCGAUGUGGGAUGCGAGGCGUCGGGCGGGCACAUGGUCGCCUGGUCGCCUGGCCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGG (((.(((((.....))))).)))((((((((((((.(((((((..((.(((((((((((((.(....).)))))))).))))).))..))...))))).).))).)))).....)))).. ( -56.90) >DroYak_CAF1 26233 112 - 1 GUUGGGCUCUCGAUGGGCAGGAUGUGAGGAUG-------UUGGCUGCGAGGCGUCGGGC-GGCACAUGGUAACAUGGUCGCUUGGUCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGG (((..((((.....))))..)))(((((....-------.....(((.(((((((((((-(((.((((....))))))))))))).))))).)))......(((.....))).))))).. ( -46.10) >consensus GUUGUGCCCUCGAUGGGCAGGAUGUGAGGAUG______GUGGGAUGCGAGGCGUCGGGCGGGCACAUGGUCACAUGGUCGCCUGGCCGCCUGGCACAUGGAUGCAAUCUGCAGUUCGCGG (((.(((((.....))))).)))...............(((((((((((((((((((((.(((.((((....))))))))))))).))))).(((......)))....)))))))))).. (-43.65 = -44.90 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:45 2006