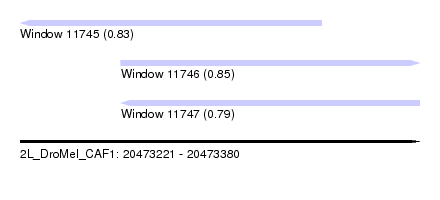
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,473,221 – 20,473,380 |
| Length | 159 |
| Max. P | 0.851537 |

| Location | 20,473,221 – 20,473,341 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.32 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20473221 120 - 22407834 AACAUUCUUGGCAAUUAACAUUUAAAAUUCCCUGCAACGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGGUGAAAUGGCAAUCGUGAAUUUUAAUUAGCAAUUUGCUGAG ......((..((((.......(((((((((..((((((((.......)))..))))).............((((((((.......)))))..))))))))))))........))))..)) ( -22.16) >DroSec_CAF1 26212 120 - 1 AACAUUCUUGGCAAUUAACAUUUAAAAUUCGCUGCAACGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGGUGAAAUGGCAAUCGUGAAUUUUAAUUAGCAAUUUGCUGGG ......((..((((.......(((((((((((((((((((.......)))..)))))................(((((.......)))))...)))))))))))........))))..)) ( -25.46) >DroSim_CAF1 28178 120 - 1 AACAUUCUUGGCAAUUAACAUUUAAAAUUCGCUGCAACGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGGUGAAAUGGCAAUCGUGAAUUUUAAUUAGCAAUUUGCUGAG ......((..((((.......(((((((((((((((((((.......)))..)))))................(((((.......)))))...)))))))))))........))))..)) ( -25.46) >DroEre_CAF1 28587 120 - 1 AACAUUCUUGGCAAUUAACAUUUAAAAUUCGCUGCAGCGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGGUGAAAUGGCAAUCGUGAAUUUUAAUUAGCAAUUUGCCGAG ......((((((((.......(((((((((((((((.(((.......)))...))))................(((((.......)))))...)))))))))))........)))))))) ( -28.06) >DroYak_CAF1 25569 120 - 1 AACAUUCUUGGCAAUUAACAUUUAAAUUUCACUGCAACGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGAUGAAAUGGCAAUCGUGAAUUUUAAUUAGCAAUUUGCUGAG ......((..((((.......(((((.(((((((((((((.......)))..)))))................(((((.......)))))...))))).)))))........))))..)) ( -22.66) >consensus AACAUUCUUGGCAAUUAACAUUUAAAAUUCGCUGCAACGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGGUGAAAUGGCAAUCGUGAAUUUUAAUUAGCAAUUUGCUGAG ......((((((((.......(((((((((((((((((((.......)))..)))))................(((((.......)))))...)))))))))))........)))))))) (-24.52 = -24.32 + -0.20)

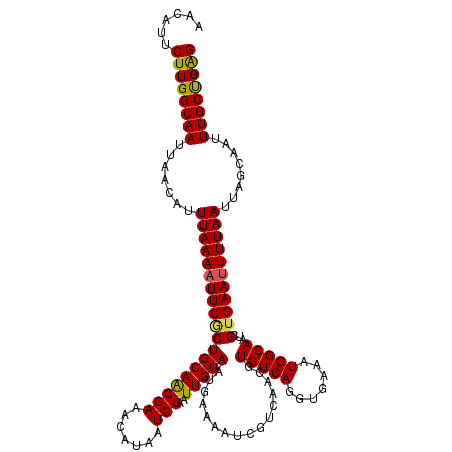

| Location | 20,473,261 – 20,473,380 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -21.12 |
| Consensus MFE | -20.14 |
| Energy contribution | -20.66 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20473261 119 + 22407834 CCUGACACGUUGACGAUUUUCAUUACAAUUCGAUUAUGUUUCGUUGCAGGGAAUUUUAAAUGUUAAUUGCCAAGAAUGUUUCAACAAAUUCAUUUCUCAUUAAUGUGUCUU-UUCAUUUA ...(((((((((((((((..((((..(((((.....(((......)))..)))))...))))..)))))....((((..........))))........))))))))))..-........ ( -20.70) >DroSec_CAF1 26252 119 + 1 CCUGACACGUUGACGAUUUUCAUUACAAUUCGAUUAUGUUUCGUUGCAGCGAAUUUUAAAUGUUAAUUGCCAAGAAUGUUUCAACAAAUUCAUUUUUCAUUAAUGUGUCUU-UUUAUUUA ...(((((((((((((((..((((..((((((....(((......))).))))))...))))..)))))..(((((((............)))))))..))))))))))..-........ ( -25.00) >DroSim_CAF1 28218 120 + 1 CCUGACACGUUGACGAUUUUCAUUACAAUUCGAUUAUGUUUCGUUGCAGCGAAUUUUAAAUGUUAAUUGCCAAGAAUGUUUCAACAAAUUCAUUUCUCAUUAAUGUGUCUUUUUUAUUUA ...(((((((((((((((..((((..((((((....(((......))).))))))...))))..)))))....((((..........))))........))))))))))........... ( -24.60) >DroEre_CAF1 28627 118 + 1 CCUGACACGUUGACGAUUUUCAUUACAAUUCGAUUAUGUUUCGCUGCAGCGAAUUUUAAAUGUUAAUUGCCAAGAAUGUUUCGACAAAUUCAUUUCUCAUUAGUGUUUCUU-UUUAU-UG ...(((((.(((.(((((..((((..((((((....(((......))).))))))...))))..))))).)))((((..........))))...........)))))....-.....-.. ( -19.90) >DroYak_CAF1 25609 118 + 1 UCUGACACGUUGACGAUUUUCAUUACAAUUCGAUUAUGUUUCGUUGCAGUGAAAUUUAAAUGUUAAUUGCCAAGAAUGUUUCAACAAAUUCAUUUCUCAUUAAUGUUUCUU-UUUAU-UA ...((.(((((((.((.(((((((.(((..(((.......)))))).)))))))...................((((..........)))).....)).))))))).))..-.....-.. ( -15.40) >consensus CCUGACACGUUGACGAUUUUCAUUACAAUUCGAUUAUGUUUCGUUGCAGCGAAUUUUAAAUGUUAAUUGCCAAGAAUGUUUCAACAAAUUCAUUUCUCAUUAAUGUGUCUU_UUUAUUUA ...(((((((((((((((..((((..((((((....(((......))).))))))...))))..)))))....((((..........))))........))))))))))........... (-20.14 = -20.66 + 0.52)

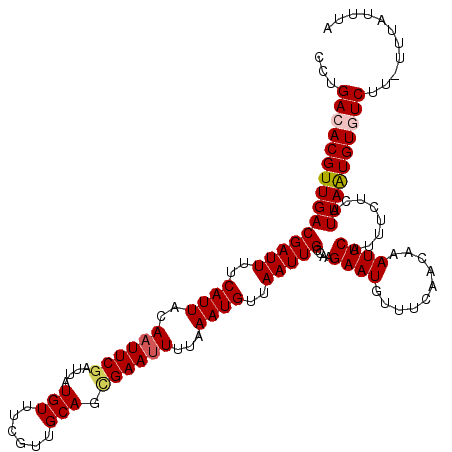
| Location | 20,473,261 – 20,473,380 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20473261 119 - 22407834 UAAAUGAA-AAGACACAUUAAUGAGAAAUGAAUUUGUUGAAACAUUCUUGGCAAUUAACAUUUAAAAUUCCCUGCAACGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGG ........-..(((((.((.((((.(((((...((((..(.......)..))))....)))))....(((..((((((((.......)))..)))))..)))..)))).)).)))))... ( -20.40) >DroSec_CAF1 26252 119 - 1 UAAAUAAA-AAGACACAUUAAUGAAAAAUGAAUUUGUUGAAACAUUCUUGGCAAUUAACAUUUAAAAUUCGCUGCAACGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGG ........-..(((((.((.((((.(((((...((((..(.......)..))))....)))))....((((.((((((((.......)))..))))).))))..)))).)).)))))... ( -21.90) >DroSim_CAF1 28218 120 - 1 UAAAUAAAAAAGACACAUUAAUGAGAAAUGAAUUUGUUGAAACAUUCUUGGCAAUUAACAUUUAAAAUUCGCUGCAACGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGG ...........(((((.((.((((.(((((...((((..(.......)..))))....)))))....((((.((((((((.......)))..))))).))))..)))).)).)))))... ( -22.00) >DroEre_CAF1 28627 118 - 1 CA-AUAAA-AAGAAACACUAAUGAGAAAUGAAUUUGUCGAAACAUUCUUGGCAAUUAACAUUUAAAAUUCGCUGCAGCGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGG ..-.....-.....((((..((((.(((((...(((((((.......)))))))....)))))....((((.((((.(((.......)))...)))).))))..))))....)))).... ( -17.20) >DroYak_CAF1 25609 118 - 1 UA-AUAAA-AAGAAACAUUAAUGAGAAAUGAAUUUGUUGAAACAUUCUUGGCAAUUAACAUUUAAAUUUCACUGCAACGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGA ..-.....-..((.((.((.((((.(((((...((((..(.......)..))))....)))))...(((((.((((((((.......)))..))))).))))).)))).)).)).))... ( -17.60) >consensus UAAAUAAA_AAGACACAUUAAUGAGAAAUGAAUUUGUUGAAACAUUCUUGGCAAUUAACAUUUAAAAUUCGCUGCAACGAAACAUAAUCGAAUUGUAAUGAAAAUCGUCAACGUGUCAGG ...........(((((.((.((((.(((((...(((((((.......)))))))....)))))....((((.((((((((.......)))..))))).))))..)))).)).)))))... (-18.64 = -19.00 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:41 2006