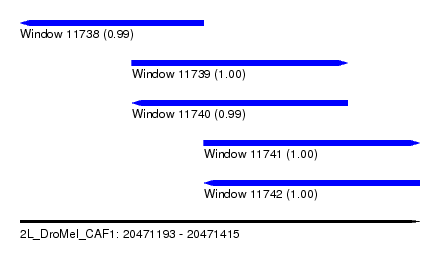
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,471,193 – 20,471,415 |
| Length | 222 |
| Max. P | 0.999795 |

| Location | 20,471,193 – 20,471,295 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -26.45 |
| Energy contribution | -26.70 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20471193 102 - 22407834 UAAUUACUUUCUAUACACACACAUUUGGCAACGGGCCAGCCGAAAUCGUUCCAUCUCCCGCCAAGUGGGAGACUCGGGCAAACAGCUCUGGUCUCUGCUAAU ........................((((((..(((((((((((..........((((((((...)))))))).))))((.....)).))))))).)))))). ( -30.40) >DroSec_CAF1 24235 87 - 1 UAAUUACUUUCUAUACACACACAUUUGGCAACGGGCC---------------AUCUCCCGCCAAGUGGGAGACUCGGGCAAACAGCUCUGGGCUCUGCUAAU ........................((((((..(((((---------------.((((((((...))))))))...((((.....))))..))))))))))). ( -30.30) >DroSim_CAF1 26174 87 - 1 UAAUUACUUUCUAUACGCACACAUUUGGCAACGGGCC---------------AUCUCCCGCCAAGUGGGAGACUCGGGCAAACAGCUCUGGUCUCUGCUAAU ................((...((((((((...(((..---------------....)))))))))))(((((((.((((.....)))).))))))))).... ( -31.80) >DroYak_CAF1 23718 102 - 1 UAAUUACUUUCUAUACACACACAUUUGGCAAUGGGCCAGCCGAAAUCGUUCCAUCUCCCGCCAAGUGGGAGACUCGGGCAAACAGCUCUGGUCUUUGCUAAU ........................(((((((.(((((((((((..........((((((((...)))))))).))))((.....)).)))))))))))))). ( -31.80) >consensus UAAUUACUUUCUAUACACACACAUUUGGCAACGGGCC_______________AUCUCCCGCCAAGUGGGAGACUCGGGCAAACAGCUCUGGUCUCUGCUAAU ........................((((((..(((((................((((((((...))))))))...((((.....)))).))))).)))))). (-26.45 = -26.70 + 0.25)
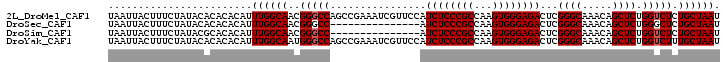
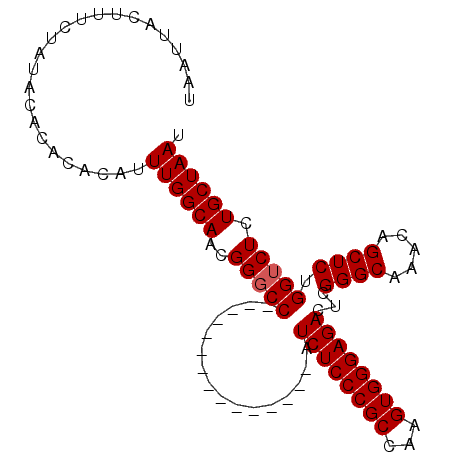
| Location | 20,471,255 – 20,471,375 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -29.58 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.999295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20471255 120 + 22407834 GCUGGCCCGUUGCCAAAUGUGUGUGUAUAGAAAGUAAUUAAGCAAUUUCAACUCGUUUUGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUGCCGAGG ((((((.....))))(((...(((((.....((....))..(((.(((((((((((..((((((..........)))))))))))))))))...))).....)))))...)))))..... ( -32.30) >DroSec_CAF1 24285 117 + 1 ---GGCCCGUUGCCAAAUGUGUGUGUAUAGAAAGUAAUUAAGCAAUUUCAACUCGUUUCGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUGCCGAGG ---(((..(((((....((((....))))....)))))...(((.((((((((((((..(((((..........)))))))))))))))))...)))................))).... ( -30.40) >DroSim_CAF1 26224 117 + 1 ---GGCCCGUUGCCAAAUGUGUGCGUAUAGAAAGUAAUUAAGCAAUUUCAACUCGUUUUGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUGCCGAGG ---(((.....)))...(((((.(((((.....((......))..(((((((((((..((((((..........)))))))))))))))))..)))))....)))))............. ( -31.40) >DroEre_CAF1 26629 120 + 1 GCUGGCCCAUUGCCAAAUGUGUGUGUAUAGAAAGUAAUUAAGCAAUUUCAACUCGUUUUGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUGCCGAGG ((((((.....))))(((...(((((.....((....))..(((.(((((((((((..((((((..........)))))))))))))))))...))).....)))))...)))))..... ( -32.30) >DroYak_CAF1 23780 120 + 1 GCUGGCCCAUUGCCAAAUGUGUGUGUAUAGAAAGUAAUUAAGCAAUUUCAACUCGUUUUGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUCCCGAGG ..((((.....))))(((...(((((.....((....))..(((.(((((((((((..((((((..........)))))))))))))))))...))).....)))))...)))....... ( -30.90) >consensus GCUGGCCCGUUGCCAAAUGUGUGUGUAUAGAAAGUAAUUAAGCAAUUUCAACUCGUUUUGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUGCCGAGG ...(((.....)))...(((((.(((((.....((......))..(((((((((((..((((((..........)))))))))))))))))..)))))....)))))............. (-29.58 = -29.62 + 0.04)

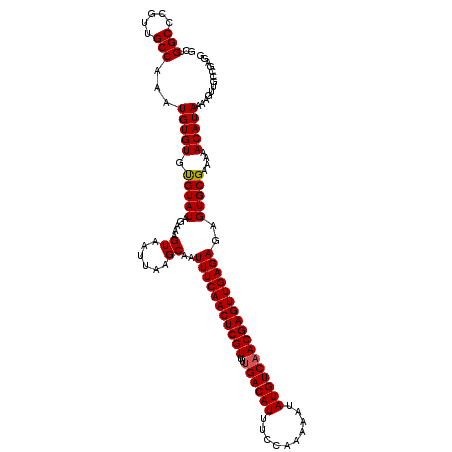
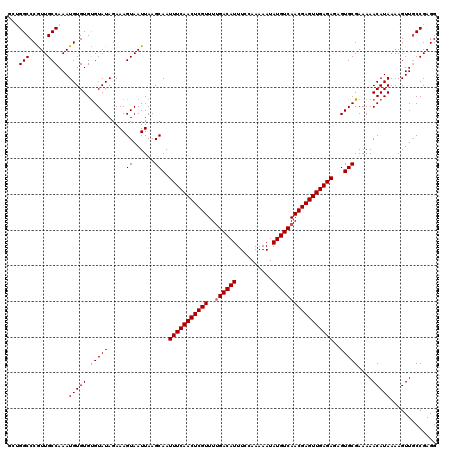
| Location | 20,471,255 – 20,471,375 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -23.84 |
| Energy contribution | -23.68 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20471255 120 - 22407834 CCUCGGCAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCAAAACGAGUUGAAAUUGCUUAAUUACUUUCUAUACACACACAUUUGGCAACGGGCCAGC ....(((................(((.....(((((((((((((((.((....)).))))))..)))))))))...))).........................(((....))))))... ( -27.20) >DroSec_CAF1 24285 117 - 1 CCUCGGCAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCGAAACGAGUUGAAAUUGCUUAAUUACUUUCUAUACACACACAUUUGGCAACGGGCC--- ....(((................(((.....(((((((((((((((.((....)).)))))..))))))))))...))).........................(((....))))))--- ( -24.80) >DroSim_CAF1 26224 117 - 1 CCUCGGCAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCAAAACGAGUUGAAAUUGCUUAAUUACUUUCUAUACGCACACAUUUGGCAACGGGCC--- .((((((((......(((.....))).....(((((((((((((((.((....)).))))))..)))))))))..)))).................((.((....))))..))))..--- ( -26.00) >DroEre_CAF1 26629 120 - 1 CCUCGGCAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCAAAACGAGUUGAAAUUGCUUAAUUACUUUCUAUACACACACAUUUGGCAAUGGGCCAGC ....(((........................(((((((((((((((.((....)).))))))..))))))))).((((((..........................))))))..)))... ( -26.97) >DroYak_CAF1 23780 120 - 1 CCUCGGGAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCAAAACGAGUUGAAAUUGCUUAAUUACUUUCUAUACACACACAUUUGGCAAUGGGCCAGC ....((((((......)))))).(((.....(((((((((((((((.((....)).))))))..)))))))))...))).........................(((((.....))))). ( -27.00) >consensus CCUCGGCAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCAAAACGAGUUGAAAUUGCUUAAUUACUUUCUAUACACACACAUUUGGCAACGGGCCAGC ....(....).............(((.....(((((((((((((((.((....)).))))))..)))))))))...)))...........................(((.....)))... (-23.84 = -23.68 + -0.16)

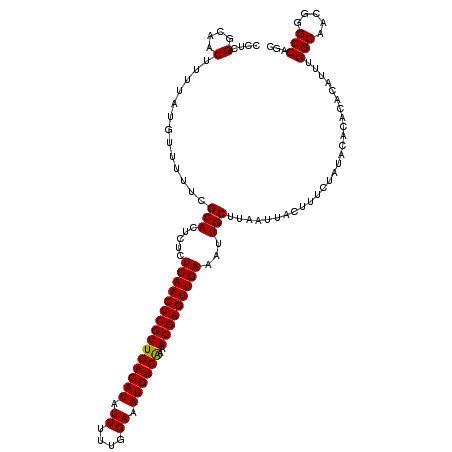
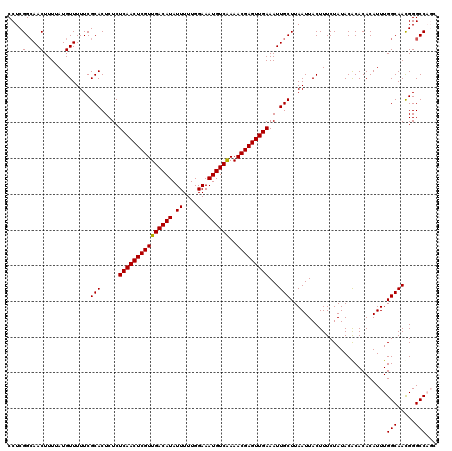
| Location | 20,471,295 – 20,471,415 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -43.28 |
| Consensus MFE | -41.26 |
| Energy contribution | -41.66 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.24 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20471295 120 + 22407834 AGCAAUUUCAACUCGUUUUGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUGCCGAGGCAAUGGCAAGCUGAUGCCGAUGGCAUUUGCCGGGCUCACA .....(((((((((((..((((((..........)))))))))))))))))((((.(.............(((((....)))))((((....((((((...))))))))))).))))... ( -43.90) >DroSec_CAF1 24322 120 + 1 AGCAAUUUCAACUCGUUUCGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUGCCGAGGCAAUGGCAAGCUGAUGCCGAUGGCAUUUGCCGGGCUCCCA .(((.((((((((((((..(((((..........)))))))))))))))))...))).............(((((....)))))((((....((((((...))))))))))((....)). ( -43.40) >DroSim_CAF1 26261 120 + 1 AGCAAUUUCAACUCGUUUUGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUGCCGAGGCAAUGGCAAGCUGAUGCCGAUGGCAUUUGCCGGGCUCCCA .(((.(((((((((((..((((((..........)))))))))))))))))...))).............(((((....)))))((((....((((((...))))))))))((....)). ( -44.10) >DroEre_CAF1 26669 120 + 1 AGCAAUUUCAACUCGUUUUGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUGCCGAGGCAAUGGCAAGCUGAUGCCGAUGGCAUUUGCCGGGCUCCCU .(((.(((((((((((..((((((..........)))))))))))))))))...))).............(((((....)))))((((....((((((...))))))))))((....)). ( -43.60) >DroYak_CAF1 23820 120 + 1 AGCAAUUUCAACUCGUUUUGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUCCCGAGGCAAUGGCAAGCUGAUGCCGAUGGCAUUUGCCGGGCUCCCU .(((.(((((((((((..((((((..........)))))))))))))))))...))).............(..(((..((((.(((....))).))))...(((....))))))..)... ( -41.40) >consensus AGCAAUUUCAACUCGUUUUGACAUUUCCAAAAAUAUGUCAACGAGUUGAGAGAGUGCGAAAAACAUAAAAGUUGCCGAGGCAAUGGCAAGCUGAUGCCGAUGGCAUUUGCCGGGCUCCCA .....(((((((((((..((((((..........)))))))))))))))))((((.(.............(((((....)))))((((....((((((...))))))))))).))))... (-41.26 = -41.66 + 0.40)

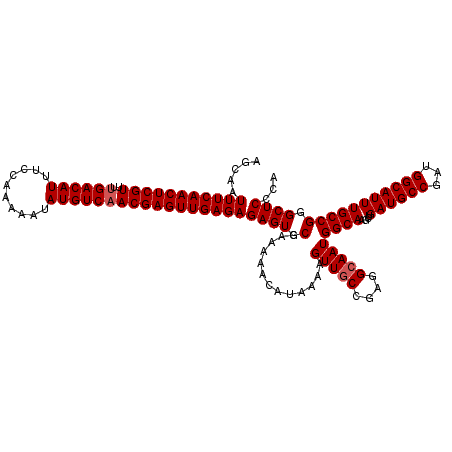
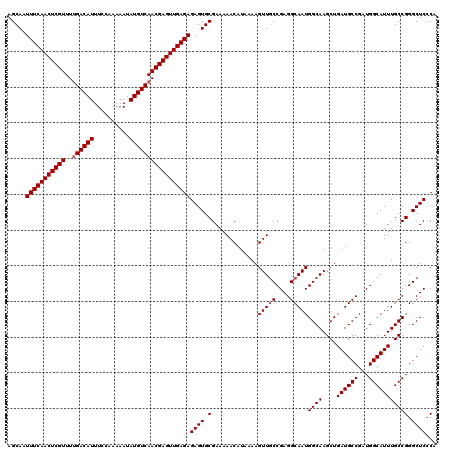
| Location | 20,471,295 – 20,471,415 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -36.68 |
| Energy contribution | -36.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20471295 120 - 22407834 UGUGAGCCCGGCAAAUGCCAUCGGCAUCAGCUUGCCAUUGCCUCGGCAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCAAAACGAGUUGAAAUUGCU ((((((((.(((....)))...((((......))))........)))(((......)))..))))).....(((((((((((((((.((....)).))))))..)))))))))....... ( -38.20) >DroSec_CAF1 24322 120 - 1 UGGGAGCCCGGCAAAUGCCAUCGGCAUCAGCUUGCCAUUGCCUCGGCAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCGAAACGAGUUGAAAUUGCU .(((((..(((((((((((...)))))....))))).((((....))))..............)..)))))(((((((((((((((.((....)).)))))..))))))))))....... ( -38.10) >DroSim_CAF1 26261 120 - 1 UGGGAGCCCGGCAAAUGCCAUCGGCAUCAGCUUGCCAUUGCCUCGGCAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCAAAACGAGUUGAAAUUGCU .(((((..(((((((((((...)))))....))))).((((....))))..............)..)))))(((((((((((((((.((....)).))))))..)))))))))....... ( -38.80) >DroEre_CAF1 26669 120 - 1 AGGGAGCCCGGCAAAUGCCAUCGGCAUCAGCUUGCCAUUGCCUCGGCAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCAAAACGAGUUGAAAUUGCU .(((((..(((((((((((...)))))....))))).((((....))))..............)..)))))(((((((((((((((.((....)).))))))..)))))))))....... ( -39.00) >DroYak_CAF1 23820 120 - 1 AGGGAGCCCGGCAAAUGCCAUCGGCAUCAGCUUGCCAUUGCCUCGGGAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCAAAACGAGUUGAAAUUGCU .(((((((((((....)))...((((......))))........)))(((......))).......)))))(((((((((((((((.((....)).))))))..)))))))))....... ( -40.00) >consensus UGGGAGCCCGGCAAAUGCCAUCGGCAUCAGCUUGCCAUUGCCUCGGCAACUUUUAUGUUUUUCGCACUCUCUCAACUCGUUGACAUAUUUUUGGAAAUGUCAAAACGAGUUGAAAUUGCU .(((((..(((((((((((...)))))....))))).((((....))))..............)..)))))(((((((((((((((.((....)).))))))..)))))))))....... (-36.68 = -36.92 + 0.24)

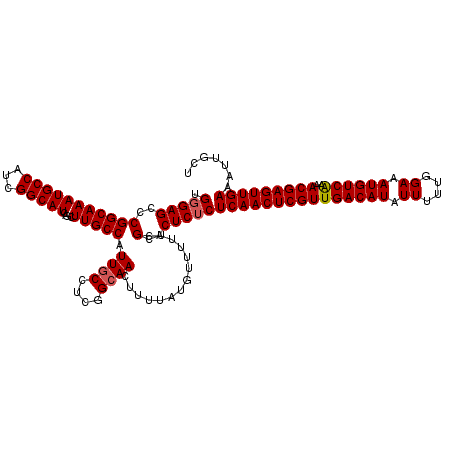
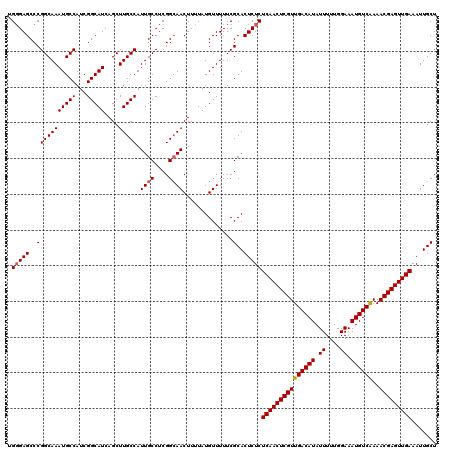
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:36 2006