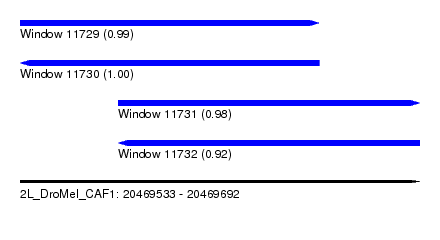
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,469,533 – 20,469,692 |
| Length | 159 |
| Max. P | 0.997994 |

| Location | 20,469,533 – 20,469,652 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.24 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -23.42 |
| Energy contribution | -22.98 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20469533 119 + 22407834 UAAAUUCCUCGAAAU-GUUUGUCACGUUCCGCACAUAAAAGGAUUUCAAUUUUCCAAUGUGACGAAAAGGCAUUUGCAUACGGAAGAAACCGUAGCAAAUAAAAUAAAAUUUUAUGUAGG ......(((((....-.(((((((((((............(((.........)))))))))))))).....((((((.(((((......)))))))))))..............)).))) ( -25.10) >DroSec_CAF1 22512 119 + 1 UAAAUUUCUUGAAAU-GUUUGUCACGUUCCGCACAUAAAAGGAUUUCAAUUUUCCAAUGUGACGAAAAGGCAUUUGCAUACGGAAGAAACCGUAGCAAAUAAAAUUAAAUUUUAUGUAGG .........((((((-.(((((((((((............(((.........)))))))))))))).....((((((.(((((......)))))))))))........))))))...... ( -25.00) >DroSim_CAF1 24464 119 + 1 UAAAUUUCUUGAAAU-GUUUGUCACGUUCCGCACAUAAAAGGAUUUCAAUUUUCCAAUGUGACGAAAAGGCAUUUGCAUACGGAAGAAACCGUAGCAAAUAAAAUCAAAUUUUAUGUAGG ........((((...-.(((((((((((............(((.........)))))))))))))).....((((((.(((((......)))))))))))....))))............ ( -25.40) >DroEre_CAF1 24777 119 + 1 UAAAUUCCUUGAAAUCGCUUGUCACGUUUCGCACAUAA-AGGAUUUCAAUUUUCCAAUGUGACAAAAAGGCAUUUGCAUACGGAAGAAACCGUAGCAAAUAAAAUCAAAUUUUAUGUUGG ........((((....((((((((((((..........-.(((.........))))))))))))....)))((((((.(((((......)))))))))))....))))............ ( -26.40) >DroYak_CAF1 21964 120 + 1 UAAAUUUCUUGAAAUCGCUUGUCACGUUUCGCACAUAAAAGGAUUUCAAUUUUCCAAUGUGACAAAAAGGCAUUUGCAUGCGGAAGAAACCGUAGCAAAUAAAAUCAAAUUUUAUCUAGG ........((((....((((((((((((............(((.........))))))))))))....)))((((((.(((((......)))))))))))....))))............ ( -26.30) >consensus UAAAUUUCUUGAAAU_GUUUGUCACGUUCCGCACAUAAAAGGAUUUCAAUUUUCCAAUGUGACGAAAAGGCAUUUGCAUACGGAAGAAACCGUAGCAAAUAAAAUCAAAUUUUAUGUAGG .........((((((.((((((((((((............(((.........)))))))))))))....))((((((.(((((......)))))))))))........))))))...... (-23.42 = -22.98 + -0.44)


| Location | 20,469,533 – 20,469,652 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.24 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -24.02 |
| Energy contribution | -23.50 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20469533 119 - 22407834 CCUACAUAAAAUUUUAUUUUAUUUGCUACGGUUUCUUCCGUAUGCAAAUGCCUUUUCGUCACAUUGGAAAAUUGAAAUCCUUUUAUGUGCGGAACGUGACAAAC-AUUUCGAGGAAUUUA ((((((((((((((((...((((((((((((......))))).)))))))..((((((......))))))..))))))...))))))).(((((.((.....))-.))))))))...... ( -27.70) >DroSec_CAF1 22512 119 - 1 CCUACAUAAAAUUUAAUUUUAUUUGCUACGGUUUCUUCCGUAUGCAAAUGCCUUUUCGUCACAUUGGAAAAUUGAAAUCCUUUUAUGUGCGGAACGUGACAAAC-AUUUCAAGAAAUUUA ........((((((.....((((((((((((......))))).)))))))...(((((.(((((.(((.........)))....))))))))))..........-........)))))). ( -24.80) >DroSim_CAF1 24464 119 - 1 CCUACAUAAAAUUUGAUUUUAUUUGCUACGGUUUCUUCCGUAUGCAAAUGCCUUUUCGUCACAUUGGAAAAUUGAAAUCCUUUUAUGUGCGGAACGUGACAAAC-AUUUCAAGAAAUUUA (((((((((((...(((((((((((((((((......))))).))))))...((((((......))))))...)))))).))))))))).))............-............... ( -26.60) >DroEre_CAF1 24777 119 - 1 CCAACAUAAAAUUUGAUUUUAUUUGCUACGGUUUCUUCCGUAUGCAAAUGCCUUUUUGUCACAUUGGAAAAUUGAAAUCCU-UUAUGUGCGAAACGUGACAAGCGAUUUCAAGGAAUUUA ((.......(((.((((..((((((((((((......))))).))))))).......)))).)))......((((((((((-((((((.....))))))..)).))))))))))...... ( -28.00) >DroYak_CAF1 21964 120 - 1 CCUAGAUAAAAUUUGAUUUUAUUUGCUACGGUUUCUUCCGCAUGCAAAUGCCUUUUUGUCACAUUGGAAAAUUGAAAUCCUUUUAUGUGCGAAACGUGACAAGCGAUUUCAAGAAAUUUA ....(((((((........(((((((..(((......)))...)))))))...)))))))...........(((((((((((((((((.....)))))).))).))))))))........ ( -27.20) >consensus CCUACAUAAAAUUUGAUUUUAUUUGCUACGGUUUCUUCCGUAUGCAAAUGCCUUUUCGUCACAUUGGAAAAUUGAAAUCCUUUUAUGUGCGGAACGUGACAAAC_AUUUCAAGAAAUUUA ........((((((.....((((((((((((......))))).)))))))...(((((.(((((.(((.........)))....))))))))))...................)))))). (-24.02 = -23.50 + -0.52)

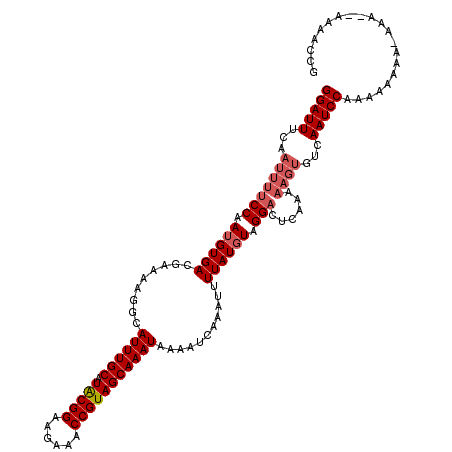
| Location | 20,469,572 – 20,469,692 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.17 |
| Mean single sequence MFE | -21.03 |
| Consensus MFE | -17.33 |
| Energy contribution | -18.37 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20469572 120 + 22407834 GGAUUUCAAUUUUCCAAUGUGACGAAAAGGCAUUUGCAUACGGAAGAAACCGUAGCAAAUAAAAUAAAAUUUUAUGUAGGACUCAAAAAGUGUCAAUCCCAAAAAAAGAAGGAAAAAUCG .........((((((................((((((.(((((......)))))))))))...................(((.(.....).)))................)))))).... ( -20.90) >DroSec_CAF1 22551 109 + 1 GGAUUUCAAUUUUCCAAUGUGACGAAAAGGCAUUUGCAUACGGAAGAAACCGUAGCAAAUAAAAUUAAAUUUUAUGUAGGAUUC----------AAUCCAAAAAAAA-AAAAAAAAACAG (((((.......(((.(((((((.....)..((((((.(((((......)))))))))))...........)))))).)))...----------)))))........-............ ( -19.70) >DroSim_CAF1 24503 118 + 1 GGAUUUCAAUUUUCCAAUGUGACGAAAAGGCAUUUGCAUACGGAAGAAACCGUAGCAAAUAAAAUCAAAUUUUAUGUAGGACUCAAAAAGUGUCAAUCCAAAAAAAA--AAAGAAAACCG ((.((((..(((((.........)))))((.((((((.(((((......)))))))))))...................(((.(.....).)))...))........--...)))).)). ( -21.30) >DroEre_CAF1 24816 117 + 1 GGAUUUCAAUUUUCCAAUGUGACAAAAAGGCAUUUGCAUACGGAAGAAACCGUAGCAAAUAAAAUCAAAUUUUAUGUUGGCCUCAAAAAGUGUGAAUCCAAAACAAA-AUA--AAAAUCC ((((((..((((((((((((((.........((((((.(((((......)))))))))))...........)))))))))......)))))..))))))........-...--....... ( -24.25) >DroYak_CAF1 22004 115 + 1 GGAUUUCAAUUUUCCAAUGUGACAAAAAGGCAUUUGCAUGCGGAAGAAACCGUAGCAAAUAAAAUCAAAUUUUAUCUAGGACUCAAAAAGUGUGAAUCCAAAAAAAA-AAG--AAAA--A (((.........)))................((((((.(((((......)))))))))))..................(((.(((.......))).)))........-...--....--. ( -19.00) >consensus GGAUUUCAAUUUUCCAAUGUGACGAAAAGGCAUUUGCAUACGGAAGAAACCGUAGCAAAUAAAAUCAAAUUUUAUGUAGGACUCAAAAAGUGUCAAUCCAAAAAAAA_AAA__AAAACCG (((((...(((((((.((((((.........((((((.(((((......)))))))))))...........)))))).)))......))))...)))))..................... (-17.33 = -18.37 + 1.04)


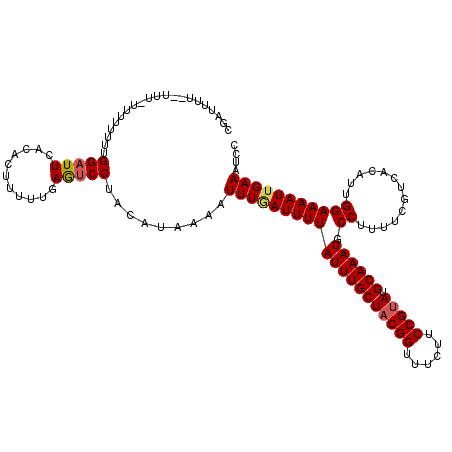
| Location | 20,469,572 – 20,469,692 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.17 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -17.18 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20469572 120 - 22407834 CGAUUUUUCCUUCUUUUUUUGGGAUUGACACUUUUUGAGUCCUACAUAAAAUUUUAUUUUAUUUGCUACGGUUUCUUCCGUAUGCAAAUGCCUUUUCGUCACAUUGGAAAAUUGAAAUCC (((((((((..........(((((((.(.......).)))))))...............((((((((((((......))))).)))))))...............)))))))))...... ( -25.50) >DroSec_CAF1 22551 109 - 1 CUGUUUUUUUUU-UUUUUUUUGGAUU----------GAAUCCUACAUAAAAUUUAAUUUUAUUUGCUACGGUUUCUUCCGUAUGCAAAUGCCUUUUCGUCACAUUGGAAAAUUGAAAUCC .....((((..(-(((((..((((((----------((((..........)))))))).((((((((((((......))))).)))))))...........))..))))))..))))... ( -20.10) >DroSim_CAF1 24503 118 - 1 CGGUUUUCUUU--UUUUUUUUGGAUUGACACUUUUUGAGUCCUACAUAAAAUUUGAUUUUAUUUGCUACGGUUUCUUCCGUAUGCAAAUGCCUUUUCGUCACAUUGGAAAAUUGAAAUCC .((.((((..(--(((((..(((((.((((..(((((.(.....).)))))..))....((((((((((((......))))).))))))).....))))).))..))))))..)))).)) ( -25.80) >DroEre_CAF1 24816 117 - 1 GGAUUUU--UAU-UUUGUUUUGGAUUCACACUUUUUGAGGCCAACAUAAAAUUUGAUUUUAUUUGCUACGGUUUCUUCCGUAUGCAAAUGCCUUUUUGUCACAUUGGAAAAUUGAAAUCC (((((((--.((-(((.(..(((((...........((((((((........))).....(((((((((((......))))).)))))))))))...))).))..).))))).))))))) ( -28.84) >DroYak_CAF1 22004 115 - 1 U--UUUU--CUU-UUUUUUUUGGAUUCACACUUUUUGAGUCCUAGAUAAAAUUUGAUUUUAUUUGCUACGGUUUCUUCCGCAUGCAAAUGCCUUUUUGUCACAUUGGAAAAUUGAAAUCC .--.(((--(..-((((((..(((((((.......)))))))..(((((((........(((((((..(((......)))...)))))))...))))))).....))))))..))))... ( -25.00) >consensus CGAUUUU__UUU_UUUUUUUUGGAUUCACACUUUUUGAGUCCUACAUAAAAUUUGAUUUUAUUUGCUACGGUUUCUUCCGUAUGCAAAUGCCUUUUCGUCACAUUGGAAAAUUGAAAUCC .....................(((((...........))))).........((((((((((((((((((((......))))).)))))).((.............))))))))))).... (-17.18 = -17.50 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:27 2006