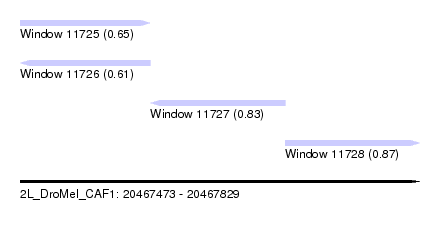
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,467,473 – 20,467,829 |
| Length | 356 |
| Max. P | 0.873127 |

| Location | 20,467,473 – 20,467,589 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -19.40 |
| Consensus MFE | -14.29 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20467473 116 + 22407834 UUUAUAGGAGAGUCAAUACGUUUCCAUAUUUUUUCCAAUACUUCGCUCACACUUUUAUGCCACACUUUAAUGUCCUGCGUGUUUUGCAGUGCAUCCGAGCCGGAAAAUAAUUUUUU ......(((((((....)).))))).....((((((........((............))....(((..(((..(((((.....)))))..)))..)))..))))))......... ( -21.90) >DroSec_CAF1 20428 116 + 1 UUAAUAAAUGAGUCAAUAAUUUUCCAUACUUUUCCCAAUACUUUUCUCACACUUUUAUGCCACACCUUUAUGCUUUGCGUUUUUCGCAGUGCAUCCGAACCGGAAAAUAAUUUUUU ..................(((((((........................((......))..........((((.(((((.....))))).)))).......)))))))........ ( -14.80) >DroSim_CAF1 22416 103 + 1 UUUAUAAAAGAGUCCAUGGUUUUCCAUAUGUUUUUCAAUACUUUGCUCACACUUUU-------------GUGCUCUGCGUUUUUCGCAGUGCAUCCGAACCGGAAAAUAAUUUUUU ...(((((((.....((((....)))).(((....(((....)))...))))))))-------------))((.(((((.....))))).)).((((...))))............ ( -21.50) >consensus UUUAUAAAAGAGUCAAUAAUUUUCCAUAUUUUUUCCAAUACUUUGCUCACACUUUUAUGCCACAC_UU_AUGCUCUGCGUUUUUCGCAGUGCAUCCGAACCGGAAAAUAAUUUUUU ..................(((((((.(((........))).............................((((.(((((.....))))).)))).......)))))))........ (-14.29 = -14.30 + 0.01)



| Location | 20,467,473 – 20,467,589 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20467473 116 - 22407834 AAAAAAUUAUUUUCCGGCUCGGAUGCACUGCAAAACACGCAGGACAUUAAAGUGUGGCAUAAAAGUGUGAGCGAAGUAUUGGAAAAAAUAUGGAAACGUAUUGACUCUCCUAUAAA .........((((((((((..((((..((((.......))))..))))..)))((.((((....))))..)).......)))))))((((((....)))))).............. ( -27.90) >DroSec_CAF1 20428 116 - 1 AAAAAAUUAUUUUCCGGUUCGGAUGCACUGCGAAAAACGCAAAGCAUAAAGGUGUGGCAUAAAAGUGUGAGAAAAGUAUUGGGAAAAGUAUGGAAAAUUAUUGACUCAUUUAUUAA ....(((.((((((((.(((.(((((..((((.....))))..((((....)))).((((....)))).......))))).)))......)))))))).))).............. ( -21.10) >DroSim_CAF1 22416 103 - 1 AAAAAAUUAUUUUCCGGUUCGGAUGCACUGCGAAAAACGCAGAGCAC-------------AAAAGUGUGAGCAAAGUAUUGAAAAACAUAUGGAAAACCAUGGACUCUUUUAUAAA ......((((.....(((((.(((((.(((((.....))))).((((-------------....)))).......))))).........((((....))))))))).....)))). ( -21.40) >consensus AAAAAAUUAUUUUCCGGUUCGGAUGCACUGCGAAAAACGCAGAGCAU_AA_GUGUGGCAUAAAAGUGUGAGCAAAGUAUUGGAAAAAAUAUGGAAAACUAUUGACUCUUUUAUAAA ..........((((((.(((.(((((.(((((.....))))).((((.................)))).......))))).)))......)))))).................... (-14.22 = -14.90 + 0.67)



| Location | 20,467,589 – 20,467,709 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -42.08 |
| Consensus MFE | -33.83 |
| Energy contribution | -35.08 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20467589 120 - 22407834 AGCUUCGGGCCAAAAAGAGCGACAGCGGCAAAAGAUGCGAAAACUUUUUCGCUGGCAGGCGUAGUAAACGUCAUUUCGCCUGGAGGCGUUUAGCGUUCGGGGGCGUGGCAAGGAGUGCAG .(((((..((((..........((((((.(((((.........)))))))))))((.(((((.....)))))......((((((.((.....)).)))))).)).))))..))))).... ( -43.30) >DroSec_CAF1 20544 120 - 1 AGCUCCGGGCCAAAAAGAGCGACAGCGGCAAAAGAUGCGAAAACUUUUUCGCUGGCAGGCGUAGUAAACGUCAUUUCGCCUGGAGGCGUUUAGCGUUCGGGGGCGUGGCAAGGAGUGCAC .(((((..((((..........((((((.(((((.........)))))))))))((.(((((.....)))))......((((((.((.....)).)))))).)).))))..))))).... ( -46.20) >DroSim_CAF1 22519 120 - 1 AGCUCCGGGCCAAAAAGAGCGACAGCGGCAAAAGAUGCGAAAACUUUUUCGCUGGCAGGCGUAGUAAACGUCAUUUCGCCUGGAGGCGUUUAGCGUUCGGGGGCGUGGCAAGGAGUGCAC .(((((..((((..........((((((.(((((.........)))))))))))((.(((((.....)))))......((((((.((.....)).)))))).)).))))..))))).... ( -46.20) >DroEre_CAF1 22964 120 - 1 AGCUCCGGGCCAAAAAGAGCGACAGCGGCAAAAGAUGCGAAAACUUUUUCGCUGGCAGGCGUAGUAAACGUCAUUUCGCCUGGAGGCGUUUAGCGUGCGGGGGCGUGGCAGGGGAUGCAC ..((((((((.(((....((..((((((.(((((.........))))))))))))).(((((.....))))).))).))))))))((((((.((.((((....))))))...)))))).. ( -42.10) >DroYak_CAF1 20246 93 - 1 AGCUCCGGGCCAAAAAGAGCGACAGCGGCAAAAGAUGCGAAAACUUUUUCGCUGGCAGGCGUAGUAAACGUCAUUUCGCCUGGAG---------------------------GAAUGCAC ..((((((((.(((....((..((((((.(((((.........))))))))))))).(((((.....))))).))).))))))))---------------------------........ ( -32.60) >consensus AGCUCCGGGCCAAAAAGAGCGACAGCGGCAAAAGAUGCGAAAACUUUUUCGCUGGCAGGCGUAGUAAACGUCAUUUCGCCUGGAGGCGUUUAGCGUUCGGGGGCGUGGCAAGGAGUGCAC ..((((((((.(((....((..((((((.(((((.........))))))))))))).(((((.....))))).))).))))))))((((((.(((((....)))))......)))))).. (-33.83 = -35.08 + 1.25)


| Location | 20,467,709 – 20,467,829 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -30.18 |
| Energy contribution | -30.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20467709 120 + 22407834 UGGGGUUUGGUCUCUGGGUCUUGGCCAAACUUUAAUGAAAAGUUUUGUUAAAAACUUUUGCUCGCUGGAAAUUUCAACGCUUGAAUGGGUCGCGGUUGGGGAUUUUUCGUGGCGGUGGUG ((((((((((((..........))))))))))))...(((((((((....)))))))))((.(((..(((((..(..(((...........)))....)..)))))..)))))....... ( -35.10) >DroSec_CAF1 20664 116 + 1 UGGGGUUUGGUCUCUGGGUCUUGGCCAAACUUUAAUGAAAAGUUUUGUUAAAAACUUUUGCUCGCUGGAAAUUUCAACGCUUGAAUGGGUCGCGGUUGGGGAUUUUCCGUCGCGG----G ((((((((((((..........))))))))))))...(((((((((....))))))))).((((((((((((..((((.(.(((.....))).)))))..)..))))))..))))----) ( -36.40) >DroSim_CAF1 22639 116 + 1 UGGGGUUUGGUCUCUGGGUCUUGGCCAAACUUUAAUGAAAAGUUUUGUUAAAAACUUUUGCUCGCUGGAAAUUUCAACGCUUGAAUGGGUCGCGGUUGGGGAUUUUCCGCGGCGG----G ((((((((((((..........))))))))))))...(((((((((....)))))))))((.(((.((((((..(..(((...........)))....)..)))))).)))))..----. ( -38.40) >DroEre_CAF1 23084 113 + 1 UGGGGUUUGGUCUCUGGGUCUUGGCCAAACUUUAAUGAAAAGUUUUGUUAAAAACUUUUGCGCGCUGGAAAUUUCAACGCUUGAAUGGGUCGCUGUUGGGGCUUUUCCGCGG-------G ((((((((((((..........))))))))))))...(((((((((....))))))))).(.(((.((((((..((((((...........)).))))..)..)))))))).-------) ( -34.90) >DroYak_CAF1 20339 113 + 1 UGAGGUUUGGUCUCUGGGUCUUGGCCAAACUUUAAUGAAAAGUUUUGUUAAAAACUUUUGCGCGCUGGAAAUUUCAACGCUUGAAUAGGUCGCUGUUGGGCAUUUUUCGGGG-------C ((((((((((((..........))))))))))))...(((((((((....)))))))))..((.(((((((.((((((((...........)).))))))...))))))).)-------) ( -34.40) >consensus UGGGGUUUGGUCUCUGGGUCUUGGCCAAACUUUAAUGAAAAGUUUUGUUAAAAACUUUUGCUCGCUGGAAAUUUCAACGCUUGAAUGGGUCGCGGUUGGGGAUUUUCCGCGGCGG____G ((((((((((((..........))))))))))))...(((((((((....)))))))))((.(((.(((((.((((((((...........)).))))))...))))))))))....... (-30.18 = -30.42 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:23 2006