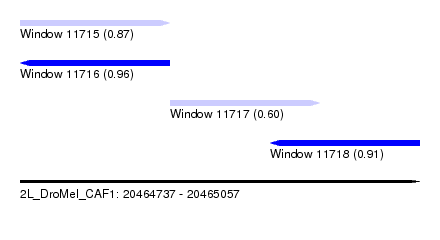
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,464,737 – 20,465,057 |
| Length | 320 |
| Max. P | 0.960766 |

| Location | 20,464,737 – 20,464,857 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -42.28 |
| Consensus MFE | -35.54 |
| Energy contribution | -35.50 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20464737 120 + 22407834 UUGUCCGCUAUUUUUAGCUGCCAACGCAGCGAUGUCGUGAGCGAAAGCGACGUUCAACGCGGGAUCGUCUUGUGAUAUUUUUAAACGACCACAUCGCUGCAGCGGCGGAGUCAAGUGCAU ...(((((((....)))).(((...((((((((((.((..((....)).........((((((.....)))))).............)).))))))))))...))))))........... ( -43.00) >DroSec_CAF1 17735 120 + 1 UUGUCCGCUAUUUUUAGCUGCCAAAGCAGCGAUGUCGUGAGCGAAAGCGACGUUCACCGCGGGAUCGUCUUGUGAUAUUUUUAAACGACCACAUCGCUGCAGCGGCGGAGUCAAGUGCAC ...(((((((....)))).(((...((((((((((.(((((((.......)))))))((((((.....))))))................))))))))))...))))))........... ( -44.60) >DroSim_CAF1 19657 120 + 1 UUGUCCGCUAUUUUUAGCUGCCAACGCAGCGAUGUCGUGAGCGAAAGCGACGUUCAACGCGGGAUCGUCUUGUGAUAUUUUUAAACGACCACAUCGCUGCAGCGGCGGAGUCAAGUGCAC ...(((((((....)))).(((...((((((((((.((..((....)).........((((((.....)))))).............)).))))))))))...))))))........... ( -43.00) >DroEre_CAF1 20170 116 + 1 UGUU--GCUAUUUUUAGCUGCCAACGCAGCGAUGUCGUGAGCGCAAACGACGGGCCACGCGGGUUAGACUUG-GAUAUUUUUAAACGACCACAUCGCUGCAGCG-CGGAGUCAAGUGCAA .((.--((((....)))).))....((((((((((((((.((.(.......).))))))..((((....(((-((....)))))..)))))))))))))).(((-(........)))).. ( -39.50) >DroYak_CAF1 17352 120 + 1 UGGUUCACUAUUUUUAGCUGCCAACGCAGCGAUGUCGUGAGCGGAAUCGACGGGGAACGCGGGAUCGUCUUGCGAUAUUUUUAAACGACCACAUCGCUGCAGCGGCGGAGUCAAGUGCAC ..((.((((...(((.(((((....((((((((((((..(((((..(((.((.....)))))..))).))..)))((....))........))))))))).))))).)))...)))).)) ( -41.30) >consensus UUGUCCGCUAUUUUUAGCUGCCAACGCAGCGAUGUCGUGAGCGAAAGCGACGUUCAACGCGGGAUCGUCUUGUGAUAUUUUUAAACGACCACAUCGCUGCAGCGGCGGAGUCAAGUGCAC ..((.((((...(((.(((((....(((((((((((((..((....)).))).....(((((((...)))))))................)))))))))).))))).)))...)))).)) (-35.54 = -35.50 + -0.04)


| Location | 20,464,737 – 20,464,857 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -40.47 |
| Consensus MFE | -33.34 |
| Energy contribution | -33.98 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20464737 120 - 22407834 AUGCACUUGACUCCGCCGCUGCAGCGAUGUGGUCGUUUAAAAAUAUCACAAGACGAUCCCGCGUUGAACGUCGCUUUCGCUCACGACAUCGCUGCGUUGGCAGCUAAAAAUAGCGGACAA ...........(((((((.(((((((((((((((((((............))))))))...(((.((.((.......)).)))))))))))))))).)))).((((....)))))))... ( -43.50) >DroSec_CAF1 17735 120 - 1 GUGCACUUGACUCCGCCGCUGCAGCGAUGUGGUCGUUUAAAAAUAUCACAAGACGAUCCCGCGGUGAACGUCGCUUUCGCUCACGACAUCGCUGCUUUGGCAGCUAAAAAUAGCGGACAA ...........(((((((..((((((((((((((((((............)))))))).....((((.((.......)).)))).))))))))))..)))).((((....)))))))... ( -44.70) >DroSim_CAF1 19657 120 - 1 GUGCACUUGACUCCGCCGCUGCAGCGAUGUGGUCGUUUAAAAAUAUCACAAGACGAUCCCGCGUUGAACGUCGCUUUCGCUCACGACAUCGCUGCGUUGGCAGCUAAAAAUAGCGGACAA ...........(((((((.(((((((((((((((((((............))))))))...(((.((.((.......)).)))))))))))))))).)))).((((....)))))))... ( -43.50) >DroEre_CAF1 20170 116 - 1 UUGCACUUGACUCCG-CGCUGCAGCGAUGUGGUCGUUUAAAAAUAUC-CAAGUCUAACCCGCGUGGCCCGUCGUUUGCGCUCACGACAUCGCUGCGUUGGCAGCUAAAAAUAGC--AACA ..(((..((((.(((-(((.(....(((.(((..(((....)))..)-)).)))....).))))))...))))..)))(((.(((.((....))))).))).((((....))))--.... ( -33.00) >DroYak_CAF1 17352 120 - 1 GUGCACUUGACUCCGCCGCUGCAGCGAUGUGGUCGUUUAAAAAUAUCGCAAGACGAUCCCGCGUUCCCCGUCGAUUCCGCUCACGACAUCGCUGCGUUGGCAGCUAAAAAUAGUGAACCA ((.((((.......((((.(((((((((((((((((((............))))))))..(((.((......))...))).....))))))))))).))))..........)))).)).. ( -37.63) >consensus GUGCACUUGACUCCGCCGCUGCAGCGAUGUGGUCGUUUAAAAAUAUCACAAGACGAUCCCGCGUUGAACGUCGCUUUCGCUCACGACAUCGCUGCGUUGGCAGCUAAAAAUAGCGGACAA ..............((((.(((((((((((((((((((............))))))..)))).......((((..........))))))))))))).)))).((((....))))...... (-33.34 = -33.98 + 0.64)

| Location | 20,464,857 – 20,464,977 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -42.65 |
| Consensus MFE | -31.18 |
| Energy contribution | -33.24 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20464857 120 + 22407834 CGAGCACACCGAGUGCAUCGAGUGCUCCGCUCUGCAUGGGCAUCAAGUGUCCGGGGGCAGUUGCAGUUGCACUGGCUGUUGCUGUUGCAUUGGCCCAACAUCGGACCAAAUGAAGCCAUU (((((((.....)))).)))(((((..(((.((((.(((((((...)))))))...))))..)).)..)))))((((..(((....)))((((.((......)).))))....))))... ( -45.80) >DroSec_CAF1 17855 114 + 1 AGAGCACACCGAGUACACCGAGUGCUCCGCUCUGCAUGGGCAUCAAGUGUCCGGGGGCAGUUGCAGUUGCA------GUUGCUGUUGCAUUGGCCCAGCAUCGGACCAAAUGAAGCCAUU (((((.....((((((.....)))))).))))).....(((.(((..(((((((((((...(((((..((.------...))..)))))...))))....))))).))..))).)))... ( -44.90) >DroSim_CAF1 19777 99 + 1 CGAGC---------ACACCGAGUGCUCCGCUCUGCAUGGGCAUCAAGUGUCCGGGGGCAGUUG------CA------GUUGCUGUUGCAUUGGCCCAGCAUCGGACCAAAUGAAGCCAUU .((((---------((.....))))))...........(((.(((..(((((((((((...((------((------(......)))))...))))....))))).))..))).)))... ( -37.80) >DroYak_CAF1 17472 105 + 1 CGAGU---------ACACCGAGUGCUCCGCGCUGCAUGGGCAUCAAGUGUCCGGGGGCAGUUGCGAUUGCA------GUUGGUGUUGCAGUGGCCCAACAUCGGACCAAAUGAAGCCAUU ...((---------(((((((.(((..((((((((.(((((((...)))))))...)))).))))...)))------.)))))).)))((((((....(((........)))..)))))) ( -42.10) >consensus CGAGC_________ACACCGAGUGCUCCGCUCUGCAUGGGCAUCAAGUGUCCGGGGGCAGUUGCAGUUGCA______GUUGCUGUUGCAUUGGCCCAACAUCGGACCAAAUGAAGCCAUU .((((((..............))))))...........(((.(((..(((((((((((...((((((.(((........))).))))))...))))....))))).))..))).)))... (-31.18 = -33.24 + 2.06)

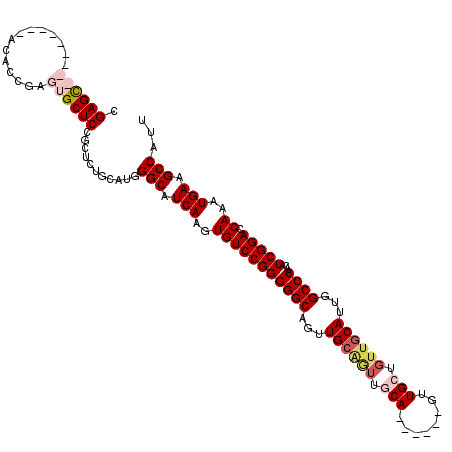
| Location | 20,464,937 – 20,465,057 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -50.10 |
| Consensus MFE | -36.22 |
| Energy contribution | -38.26 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20464937 120 - 22407834 AAUCGGGAGUGAGCAGUCCGGCGCGAGAUGCCAACUCCCGGCAAGUGUGCCUAUAUUUGGGCCACAACUCCGCGCCGACUAAUGGCUUCAUUUGGUCCGAUGUUGGGCCAAUGCAACAGC .....(((((.(..((((.((((((((.((((.......)))).(((.(((((....))))))))..)).))))))))))..).)))))..((((((((....))))))))......... ( -51.90) >DroSec_CAF1 17929 120 - 1 AAUCCGGAGUGAGGAGUCCGGCGCGAGAAGCCAACUCCCGGCGAGUGUGCCUAUAUUUGGGCCACAACUCCGCGCCGGCUAAUGGCUUCAUUUGGUCCGAUGCUGGGCCAAUGCAACAGC .....(((((.(..((.((((((((((..(((.......)))..(((.(((((....))))))))..)).))))))))))..).)))))..((((((((....))))))))......... ( -54.70) >DroSim_CAF1 19836 120 - 1 AAUCCGGAGUGAGGAGUCCGGCGCGAGAAGCCAACUCCCGGCGAGUGUGCCUAUAUUUGGGCCACAACUCCGCGACGGCUAAUGGCUUCAUUUGGUCCGAUGCUGGGCCAAUGCAACAGC ...(((((.(....).))))).(((.(((((((....((((((((((((((((....)))).))).))).)))..)))....)))))))..((((((((....)))))))))))...... ( -49.20) >DroEre_CAF1 20351 103 - 1 AAUCCGGAGCGAGGAGUCCGCCGCGAG-AGACAACUCCCGGCGAGUGUGCCUAUAUUUGGGCCACAACUCCGCGCCGGCUAAUGGCUUCAUUUGGUCCGA----------------CAAC ....((((.((((((((.(((((.(((-......))).))))).(((.(((((....)))))))).)))))..((((.....)))).....))).)))).----------------.... ( -44.80) >DroYak_CAF1 17537 120 - 1 AGUCCGGAGUGAGGAGUCCGCCGCGAGAAGACAACUCCCGGCGAGUGUGCCUAUAUUUGGGCCACAACUCCGCGCCAGCUAAUGGCUUCAUUUGGUCCGAUGUUGGGCCACUGCAACACC ........(((.(((((.(((((.(((.......))).))))).(((.(((((....)))))))).)))))((((((.....))))......(((((((....)))))))..))..))). ( -49.90) >consensus AAUCCGGAGUGAGGAGUCCGGCGCGAGAAGCCAACUCCCGGCGAGUGUGCCUAUAUUUGGGCCACAACUCCGCGCCGGCUAAUGGCUUCAUUUGGUCCGAUGCUGGGCCAAUGCAACAGC .....(((((.(..((.(((.((((((..(((.......)))..(((.(((((....))))))))..)).)))).)))))..).)))))...(((((((....))))))).......... (-36.22 = -38.26 + 2.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:14 2006