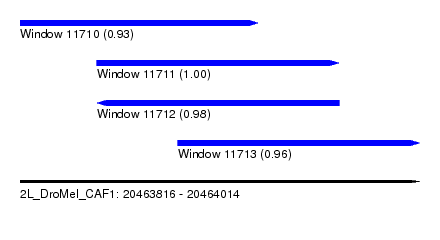
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,463,816 – 20,464,014 |
| Length | 198 |
| Max. P | 0.998777 |

| Location | 20,463,816 – 20,463,934 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -29.84 |
| Energy contribution | -31.04 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20463816 118 + 22407834 UUUAGUUGC--CGUUGCGAGCGGACUUCGCUGGGCGCAUUGUUAAUGGUCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCCGAGCCAAGCGAGGGGAAAUGGCAAAACAAACUUUCAA .....((((--(((((....)...(((((((.(((.(((.....))).(((.(((((((((((........))))))))))))))))).)))))))...))))))))............. ( -41.30) >DroSec_CAF1 16749 117 + 1 UUUAGUUGC--CGCUGCGAGUGGACUCUGCUGGGCGCAUUGUUAAUGGCCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCC-AGCCAAGCGGGGGGAAAUGGCAAAACAAACUUUCAA .....((.(--(.(((((((....))).(((((((.(((.....))))))..(((((((((((........))))))))))))-)))...)))).)).)).................... ( -37.00) >DroSim_CAF1 18671 120 + 1 UUUAGUUGCCCCGUUGCGAGUGGACUCUGCUGGGCGCAUUGUUAAUGGCCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCCGAGCCAAGCGGGGGGAAAUGGCAAAACAAACUUUCAA .....((.((((((((((..(((......)))..)))........((((((.(((((((((((........))))))))))))).))))))))))).))..................... ( -41.20) >DroEre_CAF1 19214 116 + 1 UUUAGUGUC--CG-UGCGAGUGGACUCUGCUGGGCGCAUUGUUAAUGGCCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCCUGGCCGAGCUGGAGGAAAUGGCAA-ACAAACUCUCAA ....(((((--((-.(((((....))).))))))))).(((((..((((((.(((((((((((........))))))))))).)))))).((((.......)))).)-))))........ ( -43.70) >DroYak_CAF1 16359 118 + 1 UUUAGUUGC--CGUUGAGAGUGGACUCUGCUUGGCGCAUUGUUAAUGGCCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCCAGGCCAAGCUGGAGGAAAUGGCAAAACAAACUCUCAA .........--..((((((((...(((((((((((.(((.....)))...(.(((((((((((........))))))))))))..))))))).))))....((......)).)))))))) ( -39.80) >consensus UUUAGUUGC__CGUUGCGAGUGGACUCUGCUGGGCGCAUUGUUAAUGGCCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCCGAGCCAAGCGGGGGGAAAUGGCAAAACAAACUUUCAA ...((((......((((.(.....(((((((.(((.(((.....)))...(.(((((((((((........))))))))))))..))).))))))).....).))))....))))..... (-29.84 = -31.04 + 1.20)

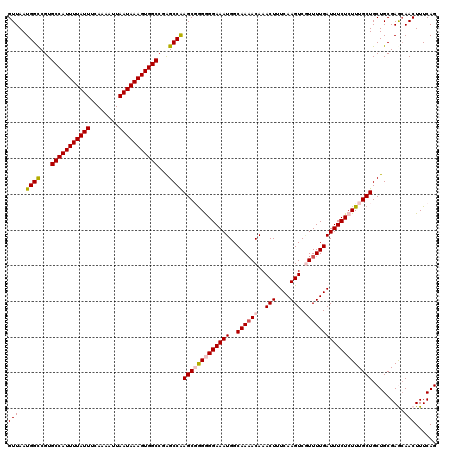
| Location | 20,463,854 – 20,463,974 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -38.66 |
| Consensus MFE | -32.02 |
| Energy contribution | -32.94 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20463854 120 + 22407834 GUUAAUGGUCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCCGAGCCAAGCGAGGGGAAAUGGCAAAACAAACUUUCAAGUCGUUUUGAUUUCUCUUUGCUGCUGCGAGCAACUUUCAG .....((((((.(((((((((((........)))))))))))))).)))(((((((((((((..((((((..(((....))).)))))))))))))))))))((.....))......... ( -40.90) >DroSec_CAF1 16787 119 + 1 GUUAAUGGCCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCC-AGCCAAGCGGGGGGAAAUGGCAAAACAAACUUUCAAGUCGUUUUGAUUUCUCUUUGCUGCUGCGAGCAACUUUCAG .....((((.(.(((((((((((........))))))))))))-.))))(((((((((((((..((((((..(((....))).)))))))))))))))))))((.....))......... ( -39.80) >DroSim_CAF1 18711 120 + 1 GUUAAUGGCCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCCGAGCCAAGCGGGGGGAAAUGGCAAAACAAACUUUCAAGUCAUUUUGAUUUCUCUUUGCUGCUGCGAGCAACUUUCAG .....((((((.(((((((((((........))))))))))))).))))(((((((((((((..(((((...(((....)))..))))))))))))))))))((.....))......... ( -39.30) >DroEre_CAF1 19251 118 + 1 GUUAAUGGCCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCCUGGCCGAGCUGGAGGAAAUGGCAA-ACAAACUCUCAAGUCUUUUUGAUUUCUCUUUGCUGCUGCGAA-AACUUUCAG .....((((((.(((((((((((........))))))))))).))))))..(((((((.........-........(((((....)))))......(((((....)))))-..))))))) ( -35.80) >DroYak_CAF1 16397 120 + 1 GUUAAUGGCCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCCAGGCCAAGCUGGAGGAAAUGGCAAAACAAACUCUCAAGUCGUUUUGAUUUCUCUUUGCUGCUGCGAACAACUUUCAG (((..(((((..(((((((((((........)))))))))))..)))))(((.(((((((....((((((..(((....))).)))))).)))))))..)))......)))......... ( -37.50) >consensus GUUAAUGGCCGUGCCAUUUUAUUUCAAAAUUAAUAAAGUGGCCGAGCCAAGCGGGGGGAAAUGGCAAAACAAACUUUCAAGUCGUUUUGAUUUCUCUUUGCUGCUGCGAGCAACUUUCAG (((..((((...(((((((((((........)))))))))))...))))(((((((((((((..((((((..(((....))).))))))))))))))))))).........)))...... (-32.02 = -32.94 + 0.92)

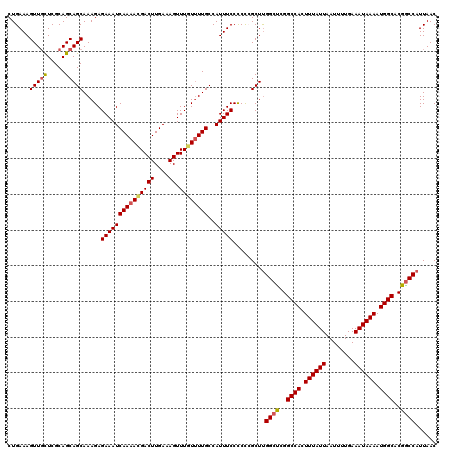
| Location | 20,463,854 – 20,463,974 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -26.26 |
| Energy contribution | -26.46 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
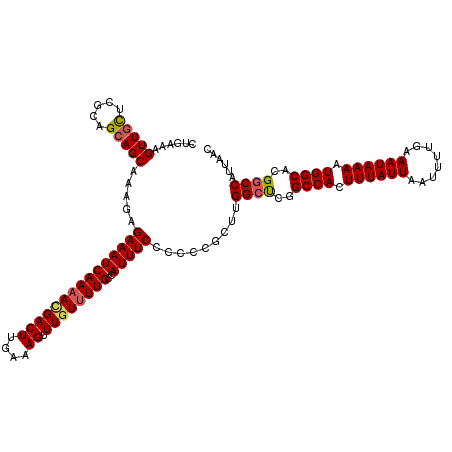
>2L_DroMel_CAF1 20463854 120 - 22407834 CUGAAAGUUGCUCGCAGCAGCAAAGAGAAAUCAAAACGACUUGAAAGUUUGUUUUGCCAUUUCCCCUCGCUUGGCUCGGCCACUUUAUUAAUUUUGAAAUAAAAUGGCACGACCAUUAAC ......(((((.....))))).....(((((((((((((((....)).))))))))..)))))........(((.(((((((.((((((........)))))).)))).))))))..... ( -30.00) >DroSec_CAF1 16787 119 - 1 CUGAAAGUUGCUCGCAGCAGCAAAGAGAAAUCAAAACGACUUGAAAGUUUGUUUUGCCAUUUCCCCCCGCUUGGCU-GGCCACUUUAUUAAUUUUGAAAUAAAAUGGCACGGCCAUUAAC ......(((((.....))))).....(((((((((((((((....)).))))))))..)))))........(((((-(((((.((((((........)))))).)))).))))))..... ( -33.70) >DroSim_CAF1 18711 120 - 1 CUGAAAGUUGCUCGCAGCAGCAAAGAGAAAUCAAAAUGACUUGAAAGUUUGUUUUGCCAUUUCCCCCCGCUUGGCUCGGCCACUUUAUUAAUUUUGAAAUAAAAUGGCACGGCCAUUAAC ......(((((.....))))).....((((((((((..(((....)).)..)))))..)))))........((((.((((((.((((((........)))))).)))).))))))..... ( -29.70) >DroEre_CAF1 19251 118 - 1 CUGAAAGUU-UUCGCAGCAGCAAAGAGAAAUCAAAAAGACUUGAGAGUUUGU-UUGCCAUUUCCUCCAGCUCGGCCAGGCCACUUUAUUAAUUUUGAAAUAAAAUGGCACGGCCAUUAAC (((...(((-.....))).(((((.(((..((((......))))...))).)-)))).........)))...((((..((((.((((((........)))))).))))..))))...... ( -29.00) >DroYak_CAF1 16397 120 - 1 CUGAAAGUUGUUCGCAGCAGCAAAGAGAAAUCAAAACGACUUGAGAGUUUGUUUUGCCAUUUCCUCCAGCUUGGCCUGGCCACUUUAUUAAUUUUGAAAUAAAAUGGCACGGCCAUUAAC (((...(((((.....)))))...(((((((((((((((((....)).))))))))..)))).))))))..(((((..((((.((((((........)))))).))))..)))))..... ( -33.00) >consensus CUGAAAGUUGCUCGCAGCAGCAAAGAGAAAUCAAAACGACUUGAAAGUUUGUUUUGCCAUUUCCCCCCGCUUGGCUCGGCCACUUUAUUAAUUUUGAAAUAAAAUGGCACGGCCAUUAAC ......(((((.....))))).....(((((((((((((((....)).))))))))..))))).........((((..((((.((((((........)))))).))))..))))...... (-26.26 = -26.46 + 0.20)

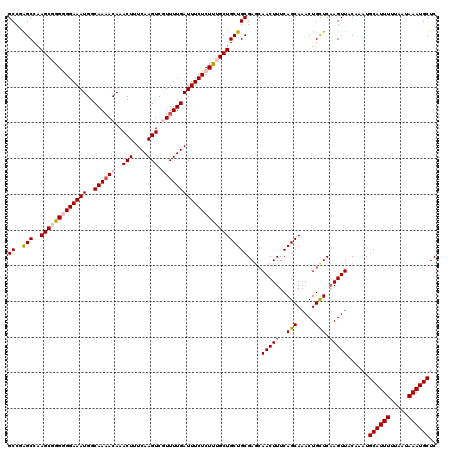
| Location | 20,463,894 – 20,464,014 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -25.76 |
| Energy contribution | -26.80 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20463894 120 + 22407834 GCCGAGCCAAGCGAGGGGAAAUGGCAAAACAAACUUUCAAGUCGUUUUGAUUUCUCUUUGCUGCUGCGAGCAACUUUCAGCAAACUGCUCAAGUUACAAAUGCAUUUUUAAUAAAUGCUC (((((((..(((((((((((((..((((((..(((....))).)))))))))))))))))))))).)).))(((((..(((.....))).)))))......((((((.....)))))).. ( -36.90) >DroSec_CAF1 16827 119 + 1 GCC-AGCCAAGCGGGGGGAAAUGGCAAAACAAACUUUCAAGUCGUUUUGAUUUCUCUUUGCUGCUGCGAGCAACUUUCAGCAAACUGCUCAAGUUACAAAUGCAUUUUUAAUAAAUGCUC ((.-(((..(((((((((((((..((((((..(((....))).))))))))))))))))))))))))(((((.............)))))...........((((((.....)))))).. ( -34.62) >DroSim_CAF1 18751 120 + 1 GCCGAGCCAAGCGGGGGGAAAUGGCAAAACAAACUUUCAAGUCAUUUUGAUUUCUCUUUGCUGCUGCGAGCAACUUUCAGCAAACUGCUCAAGUUACAAAUGCAUUUUUAAUAAAUGCUC (((((((..(((((((((((((..(((((...(((....)))..))))))))))))))))))))).)).))(((((..(((.....))).)))))......((((((.....)))))).. ( -33.20) >DroEre_CAF1 19291 117 + 1 GCCUGGCCGAGCUGGAGGAAAUGGCAA-ACAAACUCUCAAGUCUUUUUGAUUUCUCUUUGCUGCUGCGAA-AACUUUCAGCA-ACUGCUCAAGUUACAAAUGCAUUUUUAAUAAAUGCUC ((..(((..(((.(((((((.((....-.)).....(((((....))))))))))))..))))))))...-(((((..(((.-...))).)))))......((((((.....)))))).. ( -24.70) >DroYak_CAF1 16437 120 + 1 GCCAGGCCAAGCUGGAGGAAAUGGCAAAACAAACUCUCAAGUCGUUUUGAUUUCUCUUUGCUGCUGCGAACAACUUUCAGCAAACUGUUCCAGUUACAAAUGCAUUUUUAAUAAAUGCUC .((((......)))).((((((..((((((..(((....))).)))))))))))).((((..((((.(((((.............)))))))))..)))).((((((.....)))))).. ( -27.02) >consensus GCCGAGCCAAGCGGGGGGAAAUGGCAAAACAAACUUUCAAGUCGUUUUGAUUUCUCUUUGCUGCUGCGAGCAACUUUCAGCAAACUGCUCAAGUUACAAAUGCAUUUUUAAUAAAUGCUC ((..(((..(((((((((((((..((((((..(((....))).))))))))))))))))))))))))....(((((..(((.....))).)))))......((((((.....)))))).. (-25.76 = -26.80 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:09 2006