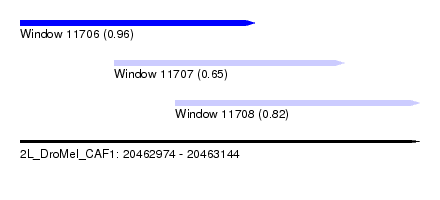
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,462,974 – 20,463,144 |
| Length | 170 |
| Max. P | 0.959304 |

| Location | 20,462,974 – 20,463,074 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -23.99 |
| Energy contribution | -25.03 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20462974 100 + 22407834 CAAUCAAUUGCCAUGACAAAUGCUCCUAGUGUUGUUGUCG--------------CUUCUGCUUGCAGCUUGGCUUGCAUUUGUCAAGUGGAAAUGCUCUAUUAUGGGGUAUUUG------ ..........(((((((((((((..((((.(((((....(--------------(....))..)))))))))...))))))))))..)))(((((((((.....))))))))).------ ( -32.60) >DroSec_CAF1 15918 100 + 1 CAAUCAAUUGCCAUGACAAAUGCUCCCAGUGUUGUUGUCG--------------CUUCUGCUUGCAGCUUGGCUUGCAUUUGUCAAGUGGAAAUGCUCUAUUAUGGGGUAUUUU------ ..........(((((((((((((..((((.(((((....(--------------(....))..)))))))))...))))))))))..)))(((((((((.....))))))))).------ ( -34.30) >DroSim_CAF1 17836 100 + 1 CAAUCAAUUGCCAUGACAAAUGCUCCCAGUGUUGUUGUCG--------------CUUCUGCUUGCAGCUUGGCUUGCAUUUGUCAAGUGGAAAUGCUCUAUUAUGGGGCAUUUU------ ..........(((((((((((((..((((.(((((....(--------------(....))..)))))))))...))))))))))..)))(((((((((.....))))))))).------ ( -36.30) >DroEre_CAF1 18406 119 + 1 CUAUCAAUUGCCAUGACAAAUGCUCCUAGUGUUGUUGUCGCCUGUUGUGUGUCUCCUCUGC-UGCAGCUUGGCUUGCAUUUGUCAAGUGGAAAUGCCCUAUUUUGCUCUAGCUUUGCUAG .............((((((((((....(((...(((((.((..(..(.....)..)...))-.)))))...))).))))))))))((..(((........)))..))(((((...))))) ( -28.10) >DroYak_CAF1 15546 116 + 1 CAAUCAAUUGCCAUGACAAAUGCUCCUAGUGUUGUUGUCGCCUGUUG----UCGCUUCUGCUUGCAGUUUGGCUUGCAUUUGUCAUGUGGAAAUGCUCUAUUUUGGUCUAGUUGCUCUAC ....(((((((((((((((((((.....(((.......)))......----..(((.(((....)))...)))..)))))))))))(((((.....)))))...)))..)))))...... ( -28.10) >consensus CAAUCAAUUGCCAUGACAAAUGCUCCUAGUGUUGUUGUCG______________CUUCUGCUUGCAGCUUGGCUUGCAUUUGUCAAGUGGAAAUGCUCUAUUAUGGGGUAUUUU______ ..........(((((((((((((..((((.(((((............................)))))))))...))))))))))..)))(((((((((.....)))))))))....... (-23.99 = -25.03 + 1.04)

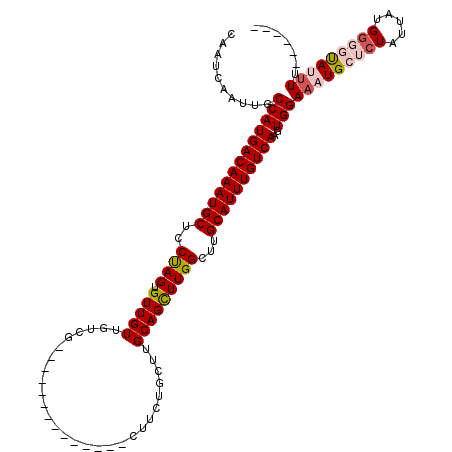
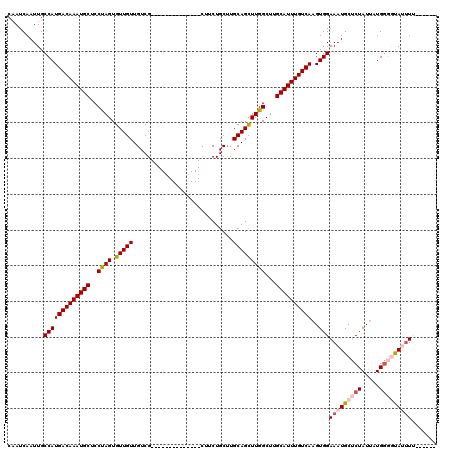
| Location | 20,463,014 – 20,463,112 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.33 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20463014 98 + 22407834 CUUCUGCUUGCAGCUUGGCUUGCAUUUGUCAAGUGGAAAUGCUCUAUUAUGGGGUAUUUGGACAUUGCUUUAAACUCGAUGGUUCUACUGGAUGGUAA ....(((..((......))..))).(..((.((((((((((((((.....))))))).....(((((.........))))).))))))).))..)... ( -23.10) >DroSec_CAF1 15958 83 + 1 CUUCUGCUUGCAGCUUGGCUUGCAUUUGUCAAGUGGAAAUGCUCUAUUAUGGGGUAUUUUGACUUUGUUU---------------UACAGGAUAGAAA .(((((((((..(((((((........))))))).((((((((((.....))))))))))..........---------------..)))).))))). ( -24.00) >DroSim_CAF1 17876 83 + 1 CUUCUGCUUGCAGCUUGGCUUGCAUUUGUCAAGUGGAAAUGCUCUAUUAUGGGGCAUUUUGACUUUGUUU---------------UACUGGAUAGAAA ....(((..((......))..)))((((((((((.((((((((((.....)))))))))).)))).....---------------.....)))))).. ( -23.30) >consensus CUUCUGCUUGCAGCUUGGCUUGCAUUUGUCAAGUGGAAAUGCUCUAUUAUGGGGUAUUUUGACUUUGUUU_______________UACUGGAUAGAAA .........((((((((((........))))))).((((((((((.....)))))))))).....))).............................. (-18.44 = -18.33 + -0.11)

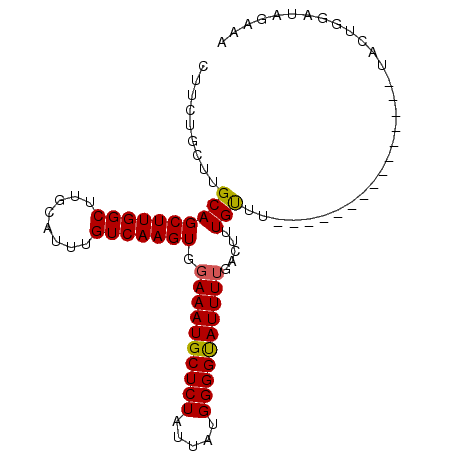
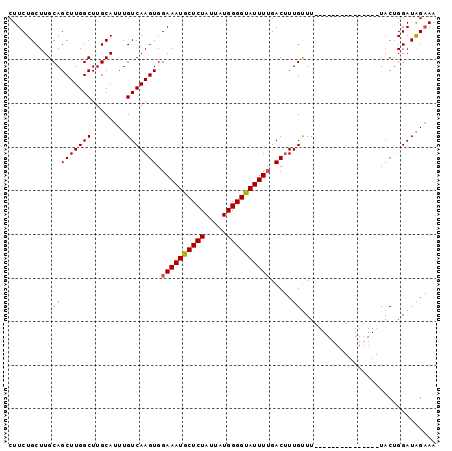
| Location | 20,463,040 – 20,463,144 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -20.83 |
| Consensus MFE | -15.16 |
| Energy contribution | -14.83 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20463040 104 + 22407834 UGUCAAGUGGAAAUGCUCUAUUAUGGGGUAUUUGGACAUUGCUUUAAACUCGAUGGUUCUACUGGAUGGUAACAUGCAAAUAUGCAAAUAUAAGAUGUAGGUAG-------- .(((.((((((((((((((.....))))))).....(((((.........))))).))))))).))).......((((....))))..................-------- ( -19.50) >DroSec_CAF1 15984 97 + 1 UGUCAAGUGGAAAUGCUCUAUUAUGGGGUAUUUUGACUUUGUUU---------------UACAGGAUAGAAACAUGUAAAUAAGCAAAUAUAAGAUACAAAUAGAGUAUAUU ............((((((((((.....((((((((..(((((((---------------(((((........).))))))...)))))..)))))))).))))))))))... ( -21.40) >DroSim_CAF1 17902 97 + 1 UGUCAAGUGGAAAUGCUCUAUUAUGGGGCAUUUUGACUUUGUUU---------------UACUGGAUAGAAACAUGUAAAUAAGCAAAUAUAAGAUAAAGAUAGAGCAUAUU ((((..((.((((((((((.....)))))))))).))(((((((---------------(((((........)).)))))...))))).....))))............... ( -21.60) >consensus UGUCAAGUGGAAAUGCUCUAUUAUGGGGUAUUUUGACUUUGUUU_______________UACUGGAUAGAAACAUGUAAAUAAGCAAAUAUAAGAUAAAGAUAGAG_AUAUU ((((..((.((((((((((.....)))))))))).)).....................................(((......))).......))))............... (-15.16 = -14.83 + -0.33)

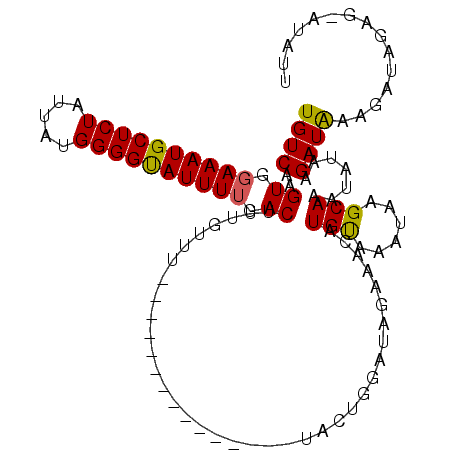

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:04 2006