
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,462,502 – 20,462,734 |
| Length | 232 |
| Max. P | 0.962151 |

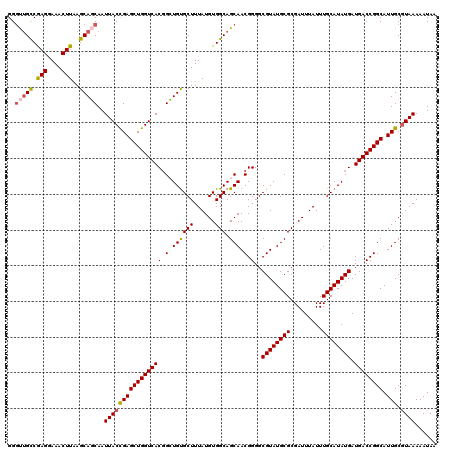
| Location | 20,462,502 – 20,462,622 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -34.20 |
| Energy contribution | -34.56 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20462502 120 + 22407834 GGGUGGUCGAGGAAACUUAAGCAGCAAUUACUGAGCUGGUCACGGCUGUGCUUUAUGUGGCAGCAACGGGGCGUAUGCGCGAUUUAUUUGCAUAUGAUGACCGGCAUUGCGUAAAAAUAA .(.((.(..((....))..).)).)..(((((((((((((((..(((((((.....)).))))).......((((((((.........)))))))).)))))))).))).))))...... ( -37.20) >DroSec_CAF1 15447 120 + 1 GGGUUGCCGAGGAAACUUAAGCAGCAAUUACCGAGCUGGUCACGGCUGUGCUUUAUGUGGCAGCAACGGGGCGUAUGCGCGAUUUAUUUGCAUAUGAUGACCGGCAUUGCGUAAAAAUAA ..(((((.(((....)))..)))))..(((((((((((((((..(((((((.....)).))))).......((((((((.........)))))))).)))))))).))).))))...... ( -42.30) >DroSim_CAF1 17365 120 + 1 GGGUUGCCGAGGAAACUUAAGCAGCAAUUACCGAGCUGGUCACGGCUGUGCUUUAUGUGGCAGCAACGGGGCGUAUGCGCGAUUUAUUUGCAUAUGAUGACCGGCAUUGCGUAAAAAUAA ..(((((.(((....)))..)))))..(((((((((((((((..(((((((.....)).))))).......((((((((.........)))))))).)))))))).))).))))...... ( -42.30) >DroEre_CAF1 17944 120 + 1 GGGCUGCCGAGCAAACUUGAGCAUCAAUUACCGAGCUGGUCACGGGUGCGCUUUAUGUGGCAGCAACGGGGCGUAUGCGCGAUUUAUUUGCAUAUGAUGACCGGCAUUGCGUAAAAAUAA .((.(((((((....)))).)))))..(((((((((((((((.(.(((((((((.(((....)))..))))))))).)((((.....))))......)))))))).))).))))...... ( -40.70) >DroYak_CAF1 15080 120 + 1 GGCUUGCCAAGAAAACUUAAGCAUCAAUUACCGAGCUGGUCACUGGUGUGCUUUAUGUGGCAGCAACGGGGCGUAUGCGCGAUUUAUUUGCAUAUGAUGACCGGCAUUGCAUAAAAAUAA .((.((((...........(((.((.......)))))(((((.(.((((((..((.((.((.((((((...))).))))).)).))...)))))).))))))))))..)).......... ( -34.20) >consensus GGGUUGCCGAGGAAACUUAAGCAGCAAUUACCGAGCUGGUCACGGCUGUGCUUUAUGUGGCAGCAACGGGGCGUAUGCGCGAUUUAUUUGCAUAUGAUGACCGGCAUUGCGUAAAAAUAA ..(((((.(((....)))..)))))..((((((((((((((((.(.((((((......))).))).).)..((((((((.........)))))))).)))))))).))).))))...... (-34.20 = -34.56 + 0.36)


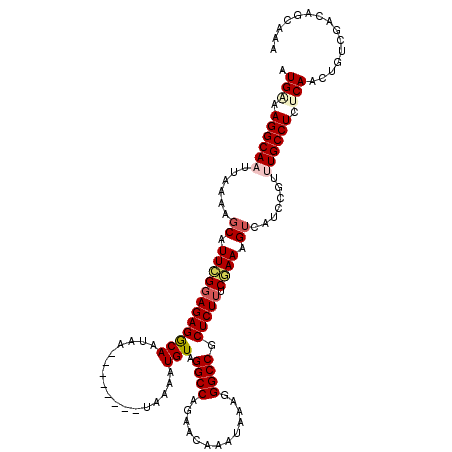
| Location | 20,462,622 – 20,462,734 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.18 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -20.57 |
| Energy contribution | -21.49 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20462622 112 + 22407834 AUGAAAGGCAAUUAAAAGCAUUCGGAGAGGCAAUAA--------UAAAAUGUAGGCCAGAACAAAUAAAGGGCCGCUCUUUCGAAAGUCAUCCGUUUGCCUUUCAACUGUCGACAGCAAA .((((((((((......((.(((((((..((.(((.--------.....))).((((.............))))))..))))))).)).......)))))))))).(((....))).... ( -32.54) >DroSec_CAF1 15567 111 + 1 AUGAAAGGCAAUUAAAAGCAUUCGGAGAGGCAAUCA--------UAAAAUGUAGGCCAGAACAAAUAAAGGGCCGCUCUUUCGAAAGUCAUCCGU-UGCCUCCCAACUGUCGACAGCAAA .((..(((((((.....((.(((((((..((...((--------(...)))..((((.............))))))..))))))).)).....))-)))))..)).(((....))).... ( -28.32) >DroSim_CAF1 17485 111 + 1 AUGAAAGGCAAUUAAAAGCAUUCGGAGAGGCAAUCG--------UAAAAUGUAGGCCAGAACAAAUAAAGGGCCGCUCUUUCGAAAGUCAUCCGU-UGCCUCCCAACUGUCGACAGCAAA .((..(((((((.....((.(((((((..((...((--------(...)))..((((.............))))))..))))))).)).....))-)))))..)).(((....))).... ( -27.92) >DroEre_CAF1 18064 119 + 1 AUGGAAGGCAAUUAAGAGCAUUCGGAGAGGCAAUAACAAAAUAACAAAAUGUAGGCCAGAACAAAU-AAGGGCCGCUCUGGCAAAAGUCAGCCGCUUGCCUCUCAGCCGUCGACAGCAAA ...((.(((......(......).(((((((((....................((((.........-...))))((.(((((....)))))..))))))))))).))).))......... ( -35.00) >DroYak_CAF1 15200 112 + 1 AUGAGAGGCAAUUAAAACCAUUUGGAGAGACAAUAA--------CAAAAUGUGGGCCAGAACAAAUGAAGGGCCGCUCUUGCAAAAGUCAGCCGUUUGCCUCUCAACAGUCGACAGCAAA .((((((((((......((....))...(((.....--------(((...((((.((............)).))))..))).....)))......))))))))))............... ( -30.30) >consensus AUGAAAGGCAAUUAAAAGCAUUCGGAGAGGCAAUAA________UAAAAUGUAGGCCAGAACAAAUAAAGGGCCGCUCUUUCGAAAGUCAUCCGUUUGCCUCUCAACUGUCGACAGCAAA .(((.((((((......((.((((((((((((.................))).((((.............)))).))))).)))).)).......)))))).)))............... (-20.57 = -21.49 + 0.92)


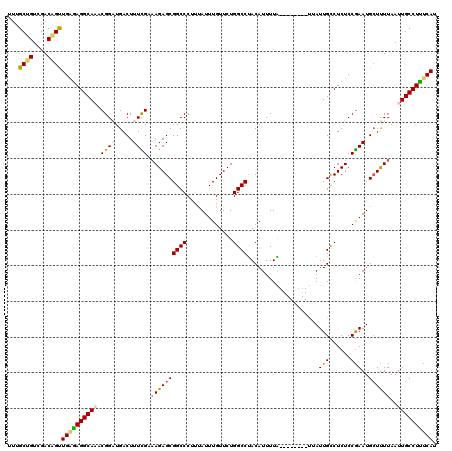
| Location | 20,462,622 – 20,462,734 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.18 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.76 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20462622 112 - 22407834 UUUGCUGUCGACAGUUGAAAGGCAAACGGAUGACUUUCGAAAGAGCGGCCCUUUAUUUGUUCUGGCCUACAUUUUA--------UUAUUGCCUCUCCGAAUGCUUUUAAUUGCCUUUCAU ...((((....))))((((((((((.((((.....)))).((((((((((.............)))).........--------...(((......)))..))))))..)))))))))). ( -29.52) >DroSec_CAF1 15567 111 - 1 UUUGCUGUCGACAGUUGGGAGGCA-ACGGAUGACUUUCGAAAGAGCGGCCCUUUAUUUGUUCUGGCCUACAUUUUA--------UGAUUGCCUCUCCGAAUGCUUUUAAUUGCCUUUCAU ...((((....)))).((((((((-(((((((...(((....))).((((.............))))..))))...--------)).))))))).))(((.((........))..))).. ( -29.92) >DroSim_CAF1 17485 111 - 1 UUUGCUGUCGACAGUUGGGAGGCA-ACGGAUGACUUUCGAAAGAGCGGCCCUUUAUUUGUUCUGGCCUACAUUUUA--------CGAUUGCCUCUCCGAAUGCUUUUAAUUGCCUUUCAU ...((((....)))).((((((((-(((((((...(((....))).((((.............))))..))))...--------)).))))))).))(((.((........))..))).. ( -31.42) >DroEre_CAF1 18064 119 - 1 UUUGCUGUCGACGGCUGAGAGGCAAGCGGCUGACUUUUGCCAGAGCGGCCCUU-AUUUGUUCUGGCCUACAUUUUGUUAUUUUGUUAUUGCCUCUCCGAAUGCUCUUAAUUGCCUUCCAU ...((((....)))).(((((((((((((.((((....((((((((((.....-..)))))))))).........))))..))))..))))))))).(((.((........)).)))... ( -36.42) >DroYak_CAF1 15200 112 - 1 UUUGCUGUCGACUGUUGAGAGGCAAACGGCUGACUUUUGCAAGAGCGGCCCUUCAUUUGUUCUGGCCCACAUUUUG--------UUAUUGUCUCUCCAAAUGGUUUUAAUUGCCUCUCAU ...............((((((((((..(((((.((((....)))))))))..(((((((....(((..((.....)--------)....)))....)))))))......)))))))))). ( -32.70) >consensus UUUGCUGUCGACAGUUGAGAGGCAAACGGAUGACUUUCGAAAGAGCGGCCCUUUAUUUGUUCUGGCCUACAUUUUA________UUAUUGCCUCUCCGAAUGCUUUUAAUUGCCUUUCAU ...((((....))))((((((((((.((((.....)))).((((((((((.............))))....................(((......)))..))))))..)))))))))). (-24.48 = -24.76 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:33:02 2006