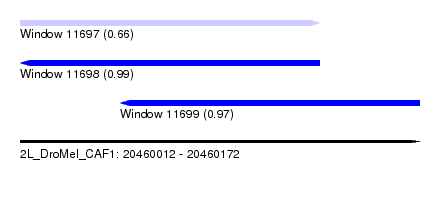
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,460,012 – 20,460,172 |
| Length | 160 |
| Max. P | 0.994072 |

| Location | 20,460,012 – 20,460,132 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.75 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
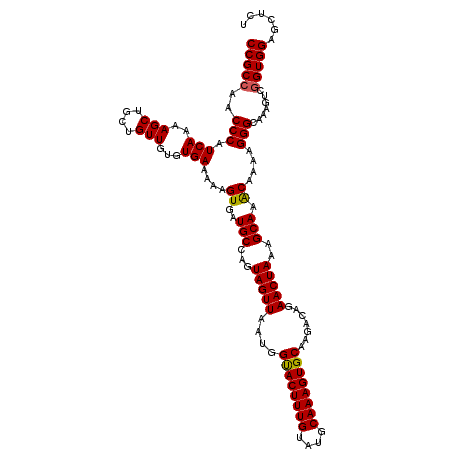
>2L_DroMel_CAF1 20460012 120 + 22407834 AUGGCCCCAUGGCUCCAUGGAUCCAUGGAUCCAGGACUCCAGAGCUCCACUGACUUUGCCCUUUUGUUUGCUUUAGUUCUGUCUUGCACUUUGCAUACAAAGUACCAUUAACUACUGGCA .(((..((((((.((....)).))))))..)))(((((....)).)))........((((...((((.(((...(((.(......).)))..))).))))((((........)))))))) ( -32.50) >DroSec_CAF1 13004 120 + 1 AUGGCCCUAUGGCCCCAUGGCUCCAUGGAACCAGGACUCCAGAGCUCCACCGACUUUGCCCCUUUGUUUGCUUUAGUUCUGUCUUGCACUUUGCAUACAAAGUACCAUUAACUACUGGCA ..((((....))))(((((....)))))..((((.....(((((.((....)))))))...((((((.(((...(((.(......).)))..))).))))))............)))).. ( -33.10) >DroSim_CAF1 14817 120 + 1 AUGGCCCGAUGGCCCCAUGGCUCCAUGGAACCAGGACUCCAGAGCUCCACCGACUUUGCCCUUUUGUUUGCUUUAGUUCUGUCUUGCACUUUGCAUACAAAGUACCAUUAACUACUGGCA ..((((....))))(((((....)))))..((((.....(((((.((....)))))))...((((((.(((...(((.(......).)))..))).))))))............)))).. ( -30.90) >DroEre_CAF1 15371 112 + 1 CUUUUCCGAUGGCUCCAGGGCUCCAGGGCUCCAGGA--------CUCCACCGACUUUGCCCUUUUGCUUGCUUUAGUUCUGUCUUGCACUUUGCAUACAAAGUGCCAUUAACUACUGGCA ......((.(((.(((.(((((....))).)).)))--------..))).))................((((.(((((..((...((((((((....)))))))).)).)))))..)))) ( -29.50) >DroYak_CAF1 13294 120 + 1 CAUUUCCGAUGGCUCCAUGGCUCCAUGGCUCUAGGACUCCAGUUCUCCACCGACUUUGCCCUUUUGUUUGCUUUAGUUCUGUCUUGCACUUUGCAUACAAAGUACCAUUAACUACUGGCA ......((.(((..(((((....)))))....(((((....)))))))).))....((((...((((.(((...(((.(......).)))..))).))))((((........)))))))) ( -26.70) >consensus AUGGCCCGAUGGCUCCAUGGCUCCAUGGAUCCAGGACUCCAGAGCUCCACCGACUUUGCCCUUUUGUUUGCUUUAGUUCUGUCUUGCACUUUGCAUACAAAGUACCAUUAACUACUGGCA .........(((..(((((....)))))..)))(((.........)))........((((...((((.(((...(((.(......).)))..))).))))((((........)))))))) (-23.02 = -23.30 + 0.28)


| Location | 20,460,012 – 20,460,132 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.75 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -31.58 |
| Energy contribution | -32.74 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20460012 120 - 22407834 UGCCAGUAGUUAAUGGUACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAACAAAAGGGCAAAGUCAGUGGAGCUCUGGAGUCCUGGAUCCAUGGAUCCAUGGAGCCAUGGGGCCAU ((((..(((((....((((((((....)))))))).......))))).............))))......((((..((((((..(((((((((....)))))).))).))).))).)))) ( -40.96) >DroSec_CAF1 13004 120 - 1 UGCCAGUAGUUAAUGGUACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAACAAAGGGGCAAAGUCGGUGGAGCUCUGGAGUCCUGGUUCCAUGGAGCCAUGGGGCCAUAGGGCCAU (((...(((((....((((((((....)))))))).......)))))..)))......(((((............))))).(.((((((((((((((....)))))))))..)))))).. ( -41.50) >DroSim_CAF1 14817 120 - 1 UGCCAGUAGUUAAUGGUACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAACAAAAGGGCAAAGUCGGUGGAGCUCUGGAGUCCUGGUUCCAUGGAGCCAUGGGGCCAUCGGGCCAU (((...(((((....((((((((....)))))))).......)))))..)))......(((((............))))).(.((((((((((((((....))))))))))..))))).. ( -41.50) >DroEre_CAF1 15371 112 - 1 UGCCAGUAGUUAAUGGCACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAGCAAAAGGGCAAAGUCGGUGGAG--------UCCUGGAGCCCUGGAGCCCUGGAGCCAUCGGAAAAG ((((..(((((....((((((((....)))))))).......)))))..(....).....))))...(((((((..--------(((.((.((......)))).))).)))))))..... ( -37.80) >DroYak_CAF1 13294 120 - 1 UGCCAGUAGUUAAUGGUACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAACAAAAGGGCAAAGUCGGUGGAGAACUGGAGUCCUAGAGCCAUGGAGCCAUGGAGCCAUCGGAAAUG ((((..(((((....((((((((....)))))))).......))))).............))))...(((((((....((((....))))..(((((....)))))..)))))))..... ( -36.16) >consensus UGCCAGUAGUUAAUGGUACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAACAAAAGGGCAAAGUCGGUGGAGCUCUGGAGUCCUGGAUCCAUGGAGCCAUGGAGCCAUCGGGCCAU (((...(((((....((((((((....)))))))).......)))))..)))......(((((....((((.......)))).)))))((..(((((....)))))..)).......... (-31.58 = -32.74 + 1.16)

| Location | 20,460,052 – 20,460,172 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -30.36 |
| Energy contribution | -30.24 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20460052 120 - 22407834 CCGCCAACCCAUCAAAAGCUGCUGUUGUGUGAAAAAGUGAUGCCAGUAGUUAAUGGUACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAACAAAAGGGCAAAGUCAGUGGAGCUCU ((((...(((.(((..(((....)))...)))....((..(((...(((((....((((((((....)))))))).......)))))..))).))....)))........))))...... ( -27.60) >DroSec_CAF1 13044 120 - 1 CCGCCAACCCAUCAAAAGCUGCUGUUGUGUGAAAAAGUGAUGCCAGUAGUUAAUGGUACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAACAAAGGGGCAAAGUCGGUGGAGCUCU (((((..(((((((..((((((((..((((.(.....).))))))))))))..)))).((((((.(((..(((.(......).)))...))).))))))))).......)))))...... ( -32.00) >DroSim_CAF1 14857 120 - 1 CCGCCAACCCAUCAAAAGCUGCUGUUGUGUGAAAAAGUGAUGCCAGUAGUUAAUGGUACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAACAAAAGGGCAAAGUCGGUGGAGCUCU (((((..(((.(((..(((....)))...)))....((..(((...(((((....((((((((....)))))))).......)))))..))).))....))).......)))))...... ( -31.10) >DroEre_CAF1 15407 116 - 1 CCGCCCACCCAUCAGAAGCUGCUGUUGUGUGAAAAAGUGAUGCCAGUAGUUAAUGGCACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAGCAAAAGGGCAAAGUCGGUGGAG---- ((((((((.((.(((......))).)).))).......(((((...(((((....((((((((....)))))))).......)))))..))..((......))...))))))))..---- ( -34.60) >DroYak_CAF1 13334 120 - 1 CCGCCAACCCAUCAGAAGCUGCUGUUGUGUGAAAAAGUGAUGCCAGUAGUUAAUGGUACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAACAAAAGGGCAAAGUCGGUGGAGAACU (((((..(((..((..((((((((..((((.(.....).))))))))))))..))((((((((....))))))))........................))).......)))))...... ( -31.40) >consensus CCGCCAACCCAUCAAAAGCUGCUGUUGUGUGAAAAAGUGAUGCCAGUAGUUAAUGGUACUUUGUAUGCAAAGUGCAAGACAGAACUAAAGCAAACAAAAGGGCAAAGUCGGUGGAGCUCU (((((..(((.(((..(((....)))...)))....((..(((...(((((....((((((((....)))))))).......)))))..))).))....))).......)))))...... (-30.36 = -30.24 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:56 2006