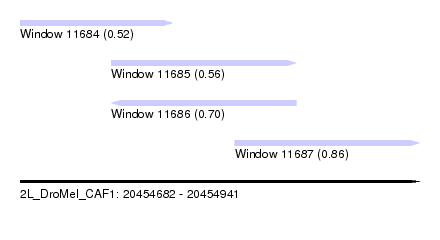
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,454,682 – 20,454,941 |
| Length | 259 |
| Max. P | 0.859988 |

| Location | 20,454,682 – 20,454,781 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -13.20 |
| Consensus MFE | -7.52 |
| Energy contribution | -7.08 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.520995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20454682 99 + 22407834 UCUAGUGUACUCUACAAACCAUCUACCAAAUUACUCUUUAAUUCUUUGAUAUCUUUUCAAUUUUCCAAAGGAGUACAAAAAUAAUUGGUUUGCACAAAU ....(((.......(((((((..........((((((((......((((.......)))).......))))))))..........)))))))))).... ( -17.38) >DroSec_CAF1 7821 84 + 1 UAUUGUGUACUCCAUAUAGCAUCUACCAAAUAACGCUUUAAUUCUUU---------------CUUCAAAAGAGUACAAAAAUAAUUGGUUUGCACUAAU ....(((((..(((((.(((..............))).))(((((((---------------.....)))))))...........)))..))))).... ( -13.24) >DroSim_CAF1 9332 84 + 1 UAUUGUGUACUCUAUAUAACAUCUACCAAAUAACGCUUUAAUUCUUU---------------CUUCAAAAGAGUACAAAAACAAUUGGUUUGCACUAAU .....((((((((..................................---------------.......))))))))......((((((....)))))) ( -8.98) >consensus UAUUGUGUACUCUAUAUAACAUCUACCAAAUAACGCUUUAAUUCUUU_______________CUUCAAAAGAGUACAAAAAUAAUUGGUUUGCACUAAU ....(((((..(((.............(((......))).(((((((....................)))))))...........)))..))))).... ( -7.52 = -7.08 + -0.44)

| Location | 20,454,741 – 20,454,861 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -20.41 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20454741 120 + 22407834 AUUUUCCAAAGGAGUACAAAAAUAAUUGGUUUGCACAAAUUUUCCGAGUGUAUUCCCAAUCCCCGCUCGCGUCCAGUAAAUUAAAUUGGUUUUUAAUGGGCCAACGAUGCGUUUUUGCAC ..........((((((((.......((((..............)))).))))))))........((.((((((..((..((((((......))))))..))....)))))).....)).. ( -23.84) >DroSec_CAF1 7868 117 + 1 ---CUUCAAAAGAGUACAAAAAUAAUUGGUUUGCACUAAUUUUUCGAGUGUAUUCCCAACCCCCGCUCGCGCCCAGUAAAUUAAAUUGGUUUUUAAUGGGCCAACGAUGCGUUUUUGCAC ---...((((((((((((.....(((((((....))))))).......)))))))........((((((.(((((..(((((.....)))))....)))))...))).))).)))))... ( -26.80) >DroSim_CAF1 9379 117 + 1 ---CUUCAAAAGAGUACAAAAACAAUUGGUUUGCACUAAUUUUUCGAGUGUAUUCCCAACCCCCGCUCGCGCCCAGUAAAUUAAAUUGGUUUUUAAUGGGCCAACGAUGCGUUUUUGCAC ---..........((.(((((((..((((..((((((.........))))))...)))).....(((((.(((((..(((((.....)))))....)))))...))).))))))))).)) ( -28.90) >DroYak_CAF1 7981 116 + 1 ---UUUUAAUAUACUAUCCAUUUAGUUGGCUUUUACUACUUUUCCAAGUGUAUUCCCAGCCGCCGU-UGCGCCCAGUAAAUUAAAUUGGUUUUUAAUGGGCCAACGAUGCGUUUUUGCUC ---.....................(((((....(((.((((....)))))))...)))))((((((-((.(((((..(((((.....)))))....))))))))))..)))......... ( -27.40) >consensus ___CUUCAAAAGAGUACAAAAAUAAUUGGUUUGCACUAAUUUUCCGAGUGUAUUCCCAACCCCCGCUCGCGCCCAGUAAAUUAAAUUGGUUUUUAAUGGGCCAACGAUGCGUUUUUGCAC ................(((((((.(((((..((((((.........))))))...)))))....(((((.(((((..(((((.....)))))....)))))...))).)))))))))... (-20.41 = -21.22 + 0.81)

| Location | 20,454,741 – 20,454,861 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -20.15 |
| Energy contribution | -23.02 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20454741 120 - 22407834 GUGCAAAAACGCAUCGUUGGCCCAUUAAAAACCAAUUUAAUUUACUGGACGCGAGCGGGGAUUGGGAAUACACUCGGAAAAUUUGUGCAAACCAAUUAUUUUUGUACUCCUUUGGAAAAU ((((((((((((.((((.(..(((.((((...........)))).))).))))))))..((((((.....(((...........)))....)))))).)))))))))(((...))).... ( -26.60) >DroSec_CAF1 7868 117 - 1 GUGCAAAAACGCAUCGUUGGCCCAUUAAAAACCAAUUUAAUUUACUGGGCGCGAGCGGGGGUUGGGAAUACACUCGAAAAAUUAGUGCAAACCAAUUAUUUUUGUACUCUUUUGAAG--- ((((((((((((.((((..(((((.((((...........)))).))))))))))))..((((((.....((((.........))))....)))))).)))))))))..........--- ( -32.40) >DroSim_CAF1 9379 117 - 1 GUGCAAAAACGCAUCGUUGGCCCAUUAAAAACCAAUUUAAUUUACUGGGCGCGAGCGGGGGUUGGGAAUACACUCGAAAAAUUAGUGCAAACCAAUUGUUUUUGUACUCUUUUGAAG--- ((((((((((((.((((..(((((.((((...........)))).)))))))))))...((((((.....((((.........))))....))))))))))))))))..........--- ( -33.80) >DroYak_CAF1 7981 116 - 1 GAGCAAAAACGCAUCGUUGGCCCAUUAAAAACCAAUUUAAUUUACUGGGCGCA-ACGGCGGCUGGGAAUACACUUGGAAAAGUAGUAAAAGCCAACUAAAUGGAUAGUAUAUUAAAA--- ..((......)).(((((((((((.((((...........)))).))))).))-)))).((((.....(((((((....)))).)))..)))).((((......)))).........--- ( -29.60) >consensus GUGCAAAAACGCAUCGUUGGCCCAUUAAAAACCAAUUUAAUUUACUGGGCGCGAGCGGGGGUUGGGAAUACACUCGAAAAAUUAGUGCAAACCAAUUAUUUUUGUACUCUUUUGAAA___ ((((((((((((.((((..(((((.((((...........)))).))))))))))))..((((((.....(((...........)))....)))))).)))))))))............. (-20.15 = -23.02 + 2.88)

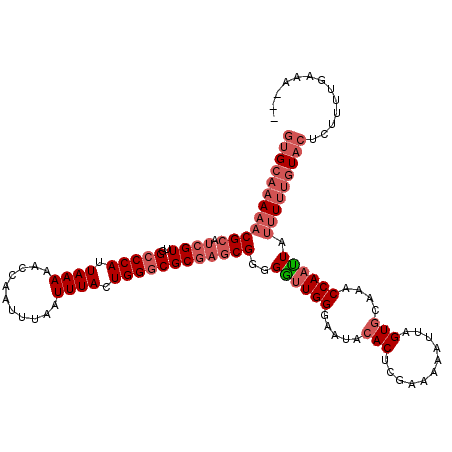
| Location | 20,454,821 – 20,454,941 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -26.70 |
| Energy contribution | -27.58 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20454821 120 + 22407834 UUAAAUUGGUUUUUAAUGGGCCAACGAUGCGUUUUUGCACAGCAUCGAUAUUGGCGCAGAUUGAUUUGGCAGUUAACAAGCAUUGCUGGCCGAAAAACAUCAAAAGUGCGCCUGGCAAAU ...................((((.((((((...........)))))).....((((((..(((((((((((((...........))).))))).....)))))...)))))))))).... ( -35.00) >DroSec_CAF1 7945 108 + 1 UUAAAUUGGUUUUUAAUGGGCCAACGAUGCGUUUUUGCACAGCAUCGAUAUUGGCGCAGAUUGAUUUGGCAGUUAACAAGCAUUGCUGGCCGAAAAACAUCAAAAGUG------------ ........((((((....(((((.(((((((((.((((.(((.((((((.(((...))))))))))))))))..)))..)))))).))))).))))))..........------------ ( -30.10) >DroSim_CAF1 9456 120 + 1 UUAAAUUGGUUUUUAAUGGGCCAACGAUGCGUUUUUGCACAGCAUCGAUAUUGGCGCAGAUUGAUUUGGCAGUUAACAAGCAUUGCUGGCCGAAAAACAUCAAAAGUGCGCCUGGGAAAU .....(((((((.....)))))))((((((...........)))))).....((((((..(((((((((((((...........))).))))).....)))))...))))))........ ( -34.00) >DroEre_CAF1 9800 115 + 1 UUAAAUUGGUUUUUAAUAGGCCAACGAUGCGUUUUUGCC---CAUCGAUAUUGGCGCAGAUUGAUUUGGCAGCUAACAAGCAUUUCUGGCC-AAAAACAUCAA-AGUGCGCCUGGGAAAU .....(((((((.....)))))))(((((.((....)).---))))).....((((((..((((((((((.(((....))).......)))-))....)))))-..))))))........ ( -33.70) >DroYak_CAF1 8057 120 + 1 UUAAAUUGGUUUUUAAUGGGCCAACGAUGCGUUUUUGCUCGGCAUCGAUAUUGGCGCAGAUUGAUUUGGCAGUUAACAAGCAUUUCUGGCCGAAAAACAUCAAAAGUUCGCCUGGCAAAU .....(((((((.....))))))).........((((((.(((.......(((((.((((.((.((((........))))))..)))))))))..(((.......))).))).)))))). ( -31.20) >consensus UUAAAUUGGUUUUUAAUGGGCCAACGAUGCGUUUUUGCACAGCAUCGAUAUUGGCGCAGAUUGAUUUGGCAGUUAACAAGCAUUGCUGGCCGAAAAACAUCAAAAGUGCGCCUGGCAAAU ........((((((....(((((.(((((((((.((((.(((.((((((.(((...))))))))))))))))..)))..)))))).))))).))))))...................... (-26.70 = -27.58 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:45 2006