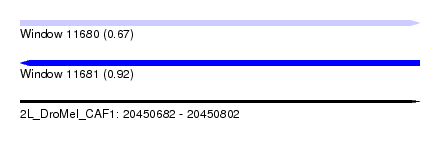
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,450,682 – 20,450,802 |
| Length | 120 |
| Max. P | 0.918836 |

| Location | 20,450,682 – 20,450,802 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -41.54 |
| Consensus MFE | -39.64 |
| Energy contribution | -39.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20450682 120 + 22407834 AAUCCGGUAUUCUGGGCGAAGAGUUCGAUGUCCCCCUCGUUGCCCGUGUAGAAGAAAAUCGGGGUGCGGGCAUUGCUCUUGUCCACGAACGAGUCGUUGUACAAGUACCGGAUGUUGAAG .((((((((((..((((.((((((..(((((((((((((.....(........).....))))).).)))))))))))))))))((((.....))))......))))))))))....... ( -43.80) >DroSec_CAF1 3668 120 + 1 AAUCCGGUAUUCUGGGCGAAGAGUUCGAUGUCCCCCUCGUUGCCCGUGUAGAAGAAGAUCGGGGUGCGGGCAUUGCUCUUGUCCACGAAUGAGUCGUUGUACAGAUACCGGAUGUUGAAG .((((((((((..((((.((((((..(((((((((((((.....(........).....))))).).)))))))))))))))))((((.....))))......))))))))))....... ( -44.20) >DroSim_CAF1 3722 120 + 1 AAUCCGGUAUUCUGGGCGAAGAGUUCGAUGUCCCCCUCGUUGCCCGUGUAGAAGAAGAUCGGGGUGCGGGCAUUGCUCUUGUCCACGAAUGAGUCGUUGUACAGAUACCGGAUGCUGAAG .((((((((((..((((.((((((..(((((((((((((.....(........).....))))).).)))))))))))))))))((((.....))))......))))))))))....... ( -44.20) >DroEre_CAF1 5925 120 + 1 AAACCGGUGUUCUGGGCGAAAAGUUCGAUGUCCCCCUCGUUGCCCGUGUAAAAGAAGAUCGGGGUGCGGUCAUUGCUCUUGUCCACGAAAGAGUCGUUGUACAAGUACCGGAUGUUGAAG ...(((((.((((((((((..((...(....)...))..)))))).......)))).)))))(((((.((((..((((((........))))))...)).))..)))))........... ( -34.01) >DroYak_CAF1 3716 120 + 1 AAUCCGGUAUUCUGGGCGAAAAGUUCGAUGUCCCCCUCGUUGCCUGUGUAAAAGAAGAUCGGGGUGCGGGCAUUGCUCUUGUCCACGAAAGAGUCGUUGUACAAAUACCGGAUGUUGAAG .((((((((((.((((((...((..((((((((((((((((..((.......))..)).))))).).))))))))..))))))))(....)............))))))))))....... ( -41.50) >consensus AAUCCGGUAUUCUGGGCGAAGAGUUCGAUGUCCCCCUCGUUGCCCGUGUAGAAGAAGAUCGGGGUGCGGGCAUUGCUCUUGUCCACGAAAGAGUCGUUGUACAAAUACCGGAUGUUGAAG .((((((((((..((((.((((((..(((((((((((((.....(........).....))))).).)))))))))))))))))((((.....))))......))))))))))....... (-39.64 = -39.64 + 0.00)



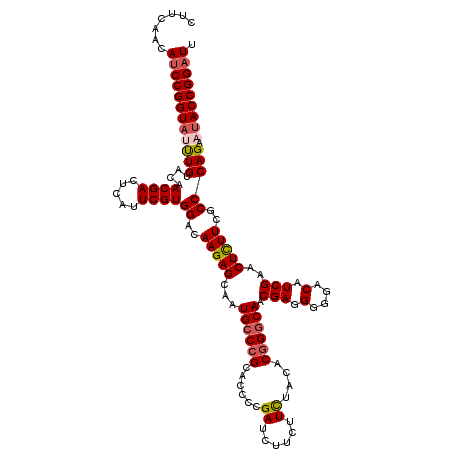
| Location | 20,450,682 – 20,450,802 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.48 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20450682 120 - 22407834 CUUCAACAUCCGGUACUUGUACAACGACUCGUUCGUGGACAAGAGCAAUGCCCGCACCCCGAUUUUCUUCUACACGGGCAACGAGGGGGACAUCGAACUCUUCGCCCAGAAUACCGGAUU .......(((((((((((((...((((.....))))..))))).((..((((((......((......))....))))))..((((((.........))))))))......)))))))). ( -38.80) >DroSec_CAF1 3668 120 - 1 CUUCAACAUCCGGUAUCUGUACAACGACUCAUUCGUGGACAAGAGCAAUGCCCGCACCCCGAUCUUCUUCUACACGGGCAACGAGGGGGACAUCGAACUCUUCGCCCAGAAUACCGGAUU .......((((((((((((....((((.....))))((..(((((...((((((......((......))....)))))).(((.(....).)))..)))))..))))).))))))))). ( -39.20) >DroSim_CAF1 3722 120 - 1 CUUCAGCAUCCGGUAUCUGUACAACGACUCAUUCGUGGACAAGAGCAAUGCCCGCACCCCGAUCUUCUUCUACACGGGCAACGAGGGGGACAUCGAACUCUUCGCCCAGAAUACCGGAUU .......((((((((((((....((((.....))))((..(((((...((((((......((......))....)))))).(((.(....).)))..)))))..))))).))))))))). ( -39.20) >DroEre_CAF1 5925 120 - 1 CUUCAACAUCCGGUACUUGUACAACGACUCUUUCGUGGACAAGAGCAAUGACCGCACCCCGAUCUUCUUUUACACGGGCAACGAGGGGGACAUCGAACUUUUCGCCCAGAACACCGGUUU .........(((((.(((((...((((.....))))..))))).((.......))....................((((...((.(....).))((.....)))))).....)))))... ( -28.90) >DroYak_CAF1 3716 120 - 1 CUUCAACAUCCGGUAUUUGUACAACGACUCUUUCGUGGACAAGAGCAAUGCCCGCACCCCGAUCUUCUUUUACACAGGCAACGAGGGGGACAUCGAACUUUUCGCCCAGAAUACCGGAUU .......(((((((((((......(((....(((((((.((.......)).)))).(((((.....).........(....)..))))......)))....)))....))))))))))). ( -31.40) >consensus CUUCAACAUCCGGUAUUUGUACAACGACUCAUUCGUGGACAAGAGCAAUGCCCGCACCCCGAUCUUCUUCUACACGGGCAACGAGGGGGACAUCGAACUCUUCGCCCAGAAUACCGGAUU .......((((((((((((....((((.....))))((..(((((...((((((......((......))....)))))).(((.(....).)))..)))))..))))).))))))))). (-31.00 = -31.48 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:40 2006