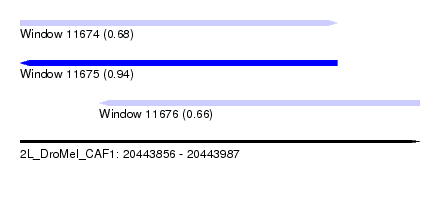
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,443,856 – 20,443,987 |
| Length | 131 |
| Max. P | 0.937616 |

| Location | 20,443,856 – 20,443,960 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -14.43 |
| Consensus MFE | -10.80 |
| Energy contribution | -10.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20443856 104 + 22407834 CAC--GCAGCAACAUGAAAUCAAAUAUAUACUAAAUAUAUAAACAUUUAUGUAUAUAUUUAGCAGACGAGUAAAUAAACCACACCCGCGCACAAAUCGCUUACAUA ...--..(((...........(((((((((((((((........))))).)))))))))).((.(.((.((...........)).))))).......)))...... ( -13.20) >DroSim_CAF1 6987 104 + 1 CAC--ACAGCAACAUGAAAUCAAAUAUAUACUAAAUAUAUAAACAUUUAUGUAUAUAUUUAGCAGACGAGUAAAUAAACCACACCCGCGCACAAAUCGCUCACAUA ...--..(((...........(((((((((((((((........))))).)))))))))).((.(.((.((...........)).))))).......)))...... ( -13.20) >DroEre_CAF1 5893 104 + 1 CACGUGCAGCAACAUAAAAUCAAAUAUAUACUAAAUAUAUAAACAUUUAUGUAUAUAUUUAGCAAACGAGUAAAUAAACCACACCCG--CACAAAUCGCUCACAUA ...(((.(((...........(((((((((((((((........))))).)))))))))).((.......................)--).......))))))... ( -16.10) >DroYak_CAF1 6932 104 + 1 CACGUGCAGCAACAUAAAAUCAAAUAUAUACUAAAUAUAUAAACAUUUAUGUAUAUAUUUAGCAAAUGAGUAAAUAAACCACACCCG--CACAAAUCGCUCUCAAA ...((((..............(((((((((((((((........))))).))))))))))......((.((......)))).....)--))).............. ( -15.20) >consensus CAC__GCAGCAACAUAAAAUCAAAUAUAUACUAAAUAUAUAAACAUUUAUGUAUAUAUUUAGCAAACGAGUAAAUAAACCACACCCG__CACAAAUCGCUCACAUA .......(((...........(((((((((((((((........))))).)))))))))).((......))..........................)))...... (-10.80 = -10.80 + -0.00)

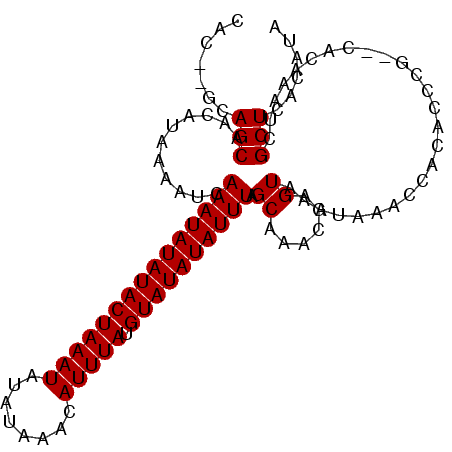
| Location | 20,443,856 – 20,443,960 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -23.58 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20443856 104 - 22407834 UAUGUAAGCGAUUUGUGCGCGGGUGUGGUUUAUUUACUCGUCUGCUAAAUAUAUACAUAAAUGUUUAUAUAUUUAGUAUAUAUUUGAUUUCAUGUUGCUGC--GUG ((((((.(((((.((...((((..(.(((......))))..))))(((((((((((.(((((((....))))))))))))))))))....)).))))))))--))) ( -30.00) >DroSim_CAF1 6987 104 - 1 UAUGUGAGCGAUUUGUGCGCGGGUGUGGUUUAUUUACUCGUCUGCUAAAUAUAUACAUAAAUGUUUAUAUAUUUAGUAUAUAUUUGAUUUCAUGUUGCUGU--GUG ......((((((.((...((((..(.(((......))))..))))(((((((((((.(((((((....))))))))))))))))))....)).))))))..--... ( -27.60) >DroEre_CAF1 5893 104 - 1 UAUGUGAGCGAUUUGUG--CGGGUGUGGUUUAUUUACUCGUUUGCUAAAUAUAUACAUAAAUGUUUAUAUAUUUAGUAUAUAUUUGAUUUUAUGUUGCUGCACGUG ((((((((((((..(((--((((((.........)))))))..))(((((((((((.(((((((....)))))))))))))))))).......)))))).)))))) ( -30.00) >DroYak_CAF1 6932 104 - 1 UUUGAGAGCGAUUUGUG--CGGGUGUGGUUUAUUUACUCAUUUGCUAAAUAUAUACAUAAAUGUUUAUAUAUUUAGUAUAUAUUUGAUUUUAUGUUGCUGCACGUG ..((..((((((....(--((((((.(((......))))))))))(((((((((((.(((((((....)))))))))))))))))).......)))))).)).... ( -26.10) >consensus UAUGUGAGCGAUUUGUG__CGGGUGUGGUUUAUUUACUCGUCUGCUAAAUAUAUACAUAAAUGUUUAUAUAUUUAGUAUAUAUUUGAUUUCAUGUUGCUGC__GUG ......((((((..(((.(((((((.........))))))).)))(((((((((((.(((((((....)))))))))))))))))).......))))))....... (-23.58 = -24.32 + 0.75)

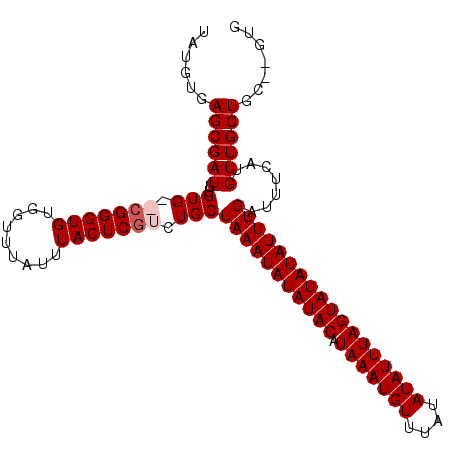
| Location | 20,443,882 – 20,443,987 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 91.79 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -18.16 |
| Energy contribution | -17.98 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
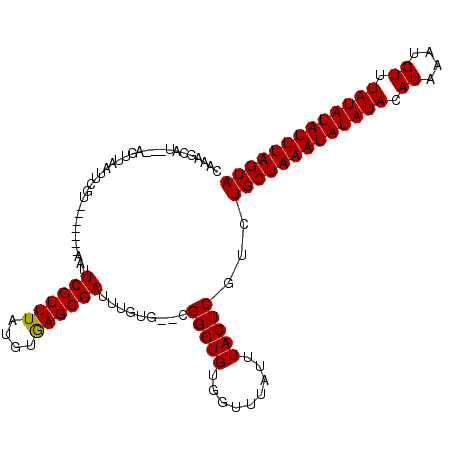
>2L_DroMel_CAF1 20443882 105 - 22407834 CAAAGCAU---AGUUAAUUCGU-------AAUUCGUUUAUGUAAGCGAUUUGUGCGCGGGUGUGGUUUAUUUACUCGUCUGCUAAAUAUAUACAUAAAUGUUUAUAUAUUUAGUA ....((((---((.........-------...((((((....)))))).))))))(((((((.........))))))).(((((((((((((.((....)).))))))))))))) ( -24.72) >DroSim_CAF1 7013 105 - 1 CAAAGCAU---AGUUAAUUCGU-------AAUUCGUUUAUGUGAGCGAUUUGUGCGCGGGUGUGGUUUAUUUACUCGUCUGCUAAAUAUAUACAUAAAUGUUUAUAUAUUUAGUA ....((((---((.........-------...((((((....)))))).))))))(((((((.........))))))).(((((((((((((.((....)).))))))))))))) ( -25.12) >DroEre_CAF1 5921 103 - 1 CAAAGCAU---AGUUAAUUCGU-------AAUUCGUUUAUGUGAGCGAUUUGUG--CGGGUGUGGUUUAUUUACUCGUUUGCUAAAUAUAUACAUAAAUGUUUAUAUAUUUAGUA ..((((..---.....((((((-------(..((((((....))))))....))--)))))((((.....))))..))))((((((((((((.((....)).)))))))))))). ( -23.50) >DroYak_CAF1 6960 113 - 1 CAAAGCAUAAUAGUUAAUUCGUAAUUGGUAAUUCGUUUUUGAGAGCGAUUUGUG--CGGGUGUGGUUUAUUUACUCAUUUGCUAAAUAUAUACAUAAAUGUUUAUAUAUUUAGUA ....((((((((((((.....)))))).....((((((....)))))).)))))--)(((((.........)))))...(((((((((((((.((....)).))))))))))))) ( -25.10) >consensus CAAAGCAU___AGUUAAUUCGU_______AAUUCGUUUAUGUGAGCGAUUUGUG__CGGGUGUGGUUUAUUUACUCGUCUGCUAAAUAUAUACAUAAAUGUUUAUAUAUUUAGUA ................................((((((....)))))).........(((((.........)))))...(((((((((((((.((....)).))))))))))))) (-18.16 = -17.98 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:35 2006