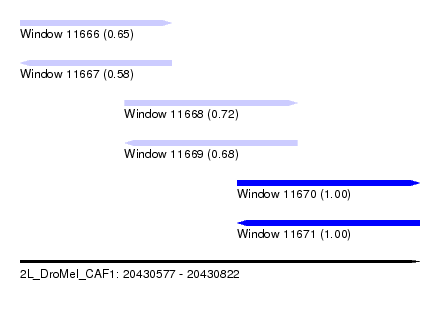
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,430,577 – 20,430,822 |
| Length | 245 |
| Max. P | 0.997546 |

| Location | 20,430,577 – 20,430,670 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.59 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -27.60 |
| Energy contribution | -27.93 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20430577 93 + 22407834 UGCCGCCACAUACCGCACUGCCAUGGUCGAGAGCAGCUCGCACGAGUUCCGGCCCUCGACCAAC------------AGC---AGCUCGAACAGCUUCGAGCUGCACGA ....((........)).......((((((((.((.((((....))))....)).))))))))..------------.((---((((((((....)))))))))).... ( -39.50) >DroVir_CAF1 12677 108 + 1 UGCCGCUACCUAUCGCACUGCCAUGGUCGAGAGCUGCUCGCACGAGUUCCGGCCCUCGACCGCCACGCCGCCAACGAGCGUGCGCUCUGACAGCUUUGAGCUGCACGA (((.(((.......((........(((((((.(((((((....))))...))).)))))))(((((((((....)).))))).)).......))....))).)))... ( -40.40) >DroPse_CAF1 9626 93 + 1 UGCCGCUACCUAUCGCACUGCCAUGGUCGAGAGCAGCUCGCACGAGUUCCGGCCCUCGACCAAC------------AGC---AGCUCAAACAGCUUUGAGCUGCACGA ....((........)).......((((((((.((.((((....))))....)).))))))))..------------.((---((((((((....)))))))))).... ( -38.20) >DroGri_CAF1 13179 105 + 1 UGCGGCUACGUAUCGCACUGCCAUGGUCGAGAGCUCAUCGCAUGAGUUUCGGCCGUCGACGACGACGCC---GACGAGCACACGCUCGGACAGCUUUGAGCUGCACGA (((((((.((.......(((((..((((((((.(((((...)))))))))))))((((.((....)).)---)))((((....)))))).)))...)))))))))... ( -42.00) >DroMoj_CAF1 15295 105 + 1 UGCCGCUACCUAUCGCACUGCCAUGGUCGAGAGCAGCUCGCACGAGUUCCGGCCCUCGACCACAACGCC---GACGAGCACACGGUCGGACAGCUUUGAGCUGCACGA (((.(((.......((.......((((((((.((.((((....))))....)).))))))))....(((---(((.........))))).).))....))).)))... ( -35.50) >DroPer_CAF1 9635 93 + 1 UGCCGCUACCUAUCGCACUGCCAUGGUCGAGAGCAGCUCGCACGAGUUCCGGCCCUCGACCAAC------------AGC---AGCUCAAACAGCUUUGAGCUGCACGA ....((........)).......((((((((.((.((((....))))....)).))))))))..------------.((---((((((((....)))))))))).... ( -38.20) >consensus UGCCGCUACCUAUCGCACUGCCAUGGUCGAGAGCAGCUCGCACGAGUUCCGGCCCUCGACCAAC____________AGC___AGCUCGAACAGCUUUGAGCUGCACGA ..............(((......((((((((.((.((((....))))....)).)))))))).....................(((((((....)))))))))).... (-27.60 = -27.93 + 0.33)

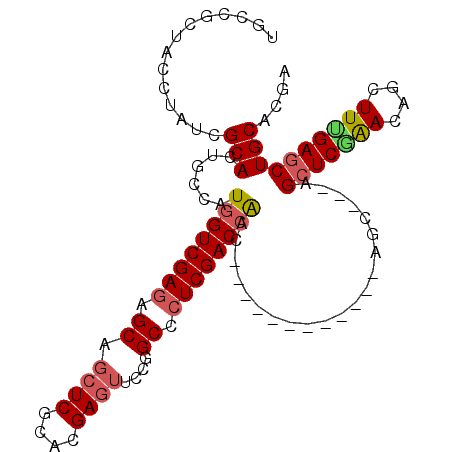
| Location | 20,430,577 – 20,430,670 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.59 |
| Mean single sequence MFE | -45.42 |
| Consensus MFE | -30.83 |
| Energy contribution | -31.67 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20430577 93 - 22407834 UCGUGCAGCUCGAAGCUGUUCGAGCU---GCU------------GUUGGUCGAGGGCCGGAACUCGUGCGAGCUGCUCUCGACCAUGGCAGUGCGGUAUGUGGCGGCA (((.(((((.....))))).)))(((---(((------------((.((((((((((.((..(((....)))))))))))))))))))))))((.((.....)).)). ( -45.90) >DroVir_CAF1 12677 108 - 1 UCGUGCAGCUCAAAGCUGUCAGAGCGCACGCUCGUUGGCGGCGUGGCGGUCGAGGGCCGGAACUCGUGCGAGCAGCUCUCGACCAUGGCAGUGCGAUAGGUAGCGGCA ...(((.((((...(((((((..((.(((((.((....)))))))))((((((((((.(...(((....)))).)))))))))).))))))).......).))).))) ( -49.20) >DroPse_CAF1 9626 93 - 1 UCGUGCAGCUCAAAGCUGUUUGAGCU---GCU------------GUUGGUCGAGGGCCGGAACUCGUGCGAGCUGCUCUCGACCAUGGCAGUGCGAUAGGUAGCGGCA (((..(((((((((....))))))))---(((------------((.((((((((((.((..(((....)))))))))))))))))))).)..)))............ ( -42.30) >DroGri_CAF1 13179 105 - 1 UCGUGCAGCUCAAAGCUGUCCGAGCGUGUGCUCGUC---GGCGUCGUCGUCGACGGCCGAAACUCAUGCGAUGAGCUCUCGACCAUGGCAGUGCGAUACGUAGCCGCA (((..((((.....))))..)))(((.(.(((((((---(((((((....)))).)))(.....)....))))))).).)......(((..((((...))))))))). ( -40.50) >DroMoj_CAF1 15295 105 - 1 UCGUGCAGCUCAAAGCUGUCCGACCGUGUGCUCGUC---GGCGUUGUGGUCGAGGGCCGGAACUCGUGCGAGCUGCUCUCGACCAUGGCAGUGCGAUAGGUAGCGGCA (((..((((.....))))..)))((((.((((..((---(((((..(((((((((((.((..(((....))))))))))))))))..)).)).)))..)))))))).. ( -52.30) >DroPer_CAF1 9635 93 - 1 UCGUGCAGCUCAAAGCUGUUUGAGCU---GCU------------GUUGGUCGAGGGCCGGAACUCGUGCGAGCUGCUCUCGACCAUGGCAGUGCGAUAGGUAGCGGCA (((..(((((((((....))))))))---(((------------((.((((((((((.((..(((....)))))))))))))))))))).)..)))............ ( -42.30) >consensus UCGUGCAGCUCAAAGCUGUCCGAGCG___GCU____________GGUGGUCGAGGGCCGGAACUCGUGCGAGCUGCUCUCGACCAUGGCAGUGCGAUAGGUAGCGGCA (((.(((((.....))))).)))......(((..............(((((((((((.....(((....)))..))))))))))).......((........))))). (-30.83 = -31.67 + 0.83)

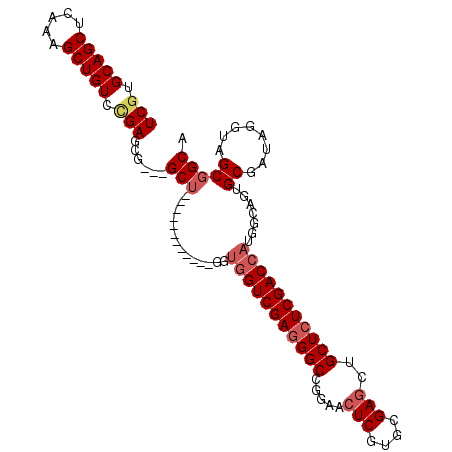
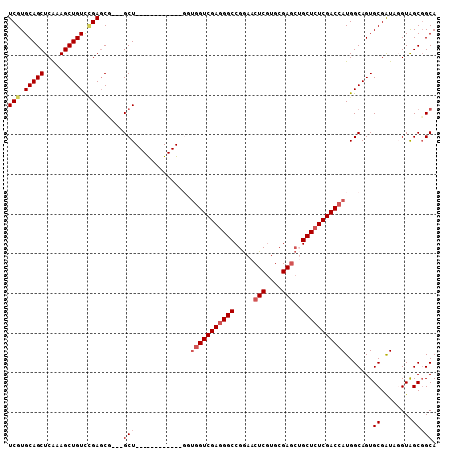
| Location | 20,430,641 – 20,430,747 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -44.19 |
| Consensus MFE | -33.73 |
| Energy contribution | -34.80 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20430641 106 + 22407834 AGCAGCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGCGUGCCGCCUCCACUGGUUUCCAGUCACCCUGGUCAUCCUGGUC .((((((((((....))))))))))..((..(((((....((((((.........((((((.....)))))).)))))).))))).))(((..((....)).))). ( -43.40) >DroSec_CAF1 2036 106 + 1 AGCAGCUCGAACAGUUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGCGUGCCGCCUCCACUGGUUGCCAGUCACCCUGGUCACCCUGGUC .((((((((((....))))))))))((.((.(((((((((((((......)))...)))))(((...))))))))..(((.(((((.....))))).))))).)). ( -47.90) >DroSim_CAF1 2970 102 + 1 AGCAGCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGCGGGCCGCCUCCACUGGUUGCCAGUCACCCUGGUCACCCU---- .((((((((((....))))))))))..(((.(((..((((((...(((........))))))))).))).)))....(((.(((((.....))))).)))..---- ( -48.50) >DroEre_CAF1 1897 106 + 1 AGCAGCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGGGCGCCGCCUUCCCUGGUUGUCAGUCAUCCUGGUCAUCUGGGUC .((((((((((....))))))))))...(((((..((......))..)))))..(((((((.....)))))))(((.(((..((((.....))))..))).))).. ( -44.70) >DroYak_CAF1 3065 106 + 1 AGCAGCUCGAACAGCUUCGAGCUGCACGAGAUGGAGGCGCCCAGUGUCAUCUGCAAGGCGGGGGUGCCGCCUCCACUGGUUGCCAGUCAUCCAGGUCAUCUAGGUC .((((((((((....)))))))))).(.(((((..(((((((..((((........)))).)))))))((((..(((((...))))).....))))))))).)... ( -48.40) >DroAna_CAF1 9030 83 + 1 AGCAGCUCGAACAGCUUCGAGCUGCACGAAAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGCCGCCGCCACCACCACC--------------UCCGGC--------- .((((((((((....)))))))))).......((.(((...(((......)))...))).))((((...........--------------..))))--------- ( -32.22) >consensus AGCAGCUCGAACAGCUUCGAGCUGCACGAGAUGGAGCCGCCCAGUGUCAUCUGCAAGGCGGGCGUGCCGCCUCCACUGGUUGCCAGUCACCCUGGUCAUCCUGGUC .((((((((((....))))))))))..(((.(((..((((((((......)))...))))).....))).)))....(((..((((.....))))..)))...... (-33.73 = -34.80 + 1.07)

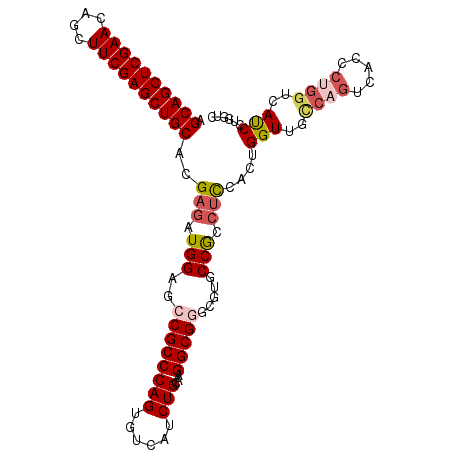
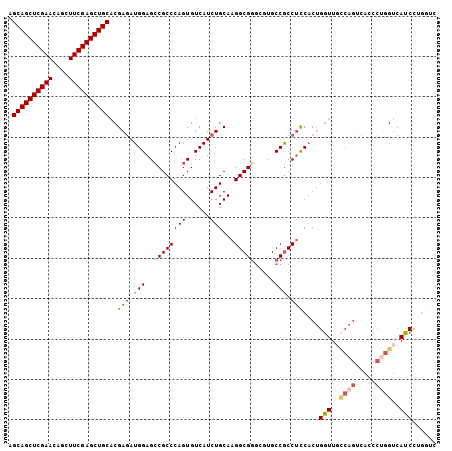
| Location | 20,430,641 – 20,430,747 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -45.53 |
| Consensus MFE | -36.08 |
| Energy contribution | -37.83 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20430641 106 - 22407834 GACCAGGAUGACCAGGGUGACUGGAAACCAGUGGAGGCGGCACGCCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGCUGCU .....(((((.((((.....))))...(((((((((((((.....))))))).(....).)))))).......)))))...((((((((((....)))))))))). ( -47.10) >DroSec_CAF1 2036 106 - 1 GACCAGGGUGACCAGGGUGACUGGCAACCAGUGGAGGCGGCACGCCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAACUGUUCGAGCUGCU .....((....)).(((((...(((..(((((((((((((.....))))))).(....).))))))...))).)))))...((((((((((....)))))))))). ( -49.10) >DroSim_CAF1 2970 102 - 1 ----AGGGUGACCAGGGUGACUGGCAACCAGUGGAGGCGGCCCGCCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGCUGCU ----.((....)).(((((...(((..(((((((((((((.....))))))).(....).))))))...))).)))))...((((((((((....)))))))))). ( -49.00) >DroEre_CAF1 1897 106 - 1 GACCCAGAUGACCAGGAUGACUGACAACCAGGGAAGGCGGCGCCCCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGCUGCU (.(((((.((..(((.....))).....((.(.(((((((.....))))))).)...)).)))))))).............((((((((((....)))))))))). ( -42.80) >DroYak_CAF1 3065 106 - 1 GACCUAGAUGACCUGGAUGACUGGCAACCAGUGGAGGCGGCACCCCCGCCUUGCAGAUGACACUGGGCGCCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGCUGCU .......((((..((((.....(((..(((((((((((((.....))))))).(....).))))))..)))))))..))))((((((((((....)))))))))). ( -48.50) >DroAna_CAF1 9030 83 - 1 ---------GCCGGA--------------GGUGGUGGUGGCGGCGGCGCCUUGCAGAUGACACUGGGCGGCUCCAUUUCGUGCAGCUCGAAGCUGUUCGAGCUGCU ---------....((--------------(((((.(((.(((((...)))...(((......))).)).))))))))))..((((((((((....)))))))))). ( -36.70) >consensus GACCAGGAUGACCAGGGUGACUGGCAACCAGUGGAGGCGGCACCCCCGCCUUGCAGAUGACACUGGGCGGCUCCAUCUCGUGCAGCUCGAAGCUGUUCGAGCUGCU .....(((((.((((.....))))...(((((((((((((.....))))))).(....).)))))).......)))))...((((((((((....)))))))))). (-36.08 = -37.83 + 1.75)
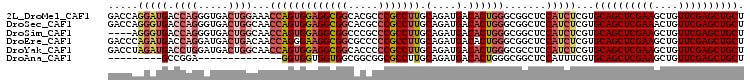

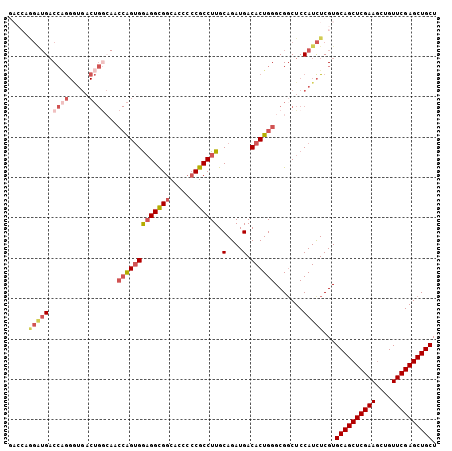
| Location | 20,430,710 – 20,430,822 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -48.38 |
| Consensus MFE | -37.27 |
| Energy contribution | -41.24 |
| Covariance contribution | 3.97 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20430710 112 + 22407834 CCUCCACUGGUUUCCAGUCACCCUGGUCAUCCUGGUCAUCCUGAUCAUCCGCCGCCGCGGUACCAAUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGA .((((((((((.....((((.((.((((....(((((.....)))))....(((((..(((((........))))).))))).)))).)).)))).))))))))))...... ( -52.10) >DroSec_CAF1 2105 112 + 1 CCUCCACUGGUUGCCAGUCACCCUGGUCACCCUGGUCAUCCUGGUCAUCCGCCGCCGCGGUACCAGUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGA .((((((((((.(((((.....)))))......(((((.((.((((.((((((.....(((((........))))).)))))))))).)).)))))))))))))))...... ( -58.30) >DroSim_CAF1 3039 104 + 1 CCUCCACUGGUUGCCAGUCACCCUGGUCACCCU--------UGAAGACGCGCCGCCGCGGUACCAAUUCGAGUACCAGGCGGAGACCCGGNNNNNNNNNNNNNNNNNNNNNN .((.((..(((.(((((.....))))).)))..--------)).)).((..(((((..(((((........))))).))))).....))....................... ( -29.70) >DroEre_CAF1 1966 112 + 1 CCUUCCCUGGUUGUCAGUCAUCCUGGUCAUCUGGGUCAUCCUGGUCAUCCGCCGCCGCGGUACCAAUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGA ..(((((((((.(((((((..((..(....)..)).......((((.((((((.....(((((........))))).)))))))))).))))))).))))).))))...... ( -51.20) >DroYak_CAF1 3134 112 + 1 CCUCCACUGGUUGCCAGUCAUCCAGGUCAUCUAGGUCAUCCAGGUCAUCCGCCGCCGCGAUACCAAUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGA .((((((((((.(((.........)))......(((((.((.((((.((((((....(((.......))).......)))))))))).)).)))))))))))))))...... ( -50.60) >consensus CCUCCACUGGUUGCCAGUCACCCUGGUCAUCCUGGUCAUCCUGGUCAUCCGCCGCCGCGGUACCAAUUCGAGUACCAGGCGGAGACCCGGCUGACCACCAGUGGGGUGCAGA .((((((((((.(((((.....)))))......(((((.((.((((.....(((((..(((((........))))).))))).)))).)).)))))))))))))))...... (-37.27 = -41.24 + 3.97)

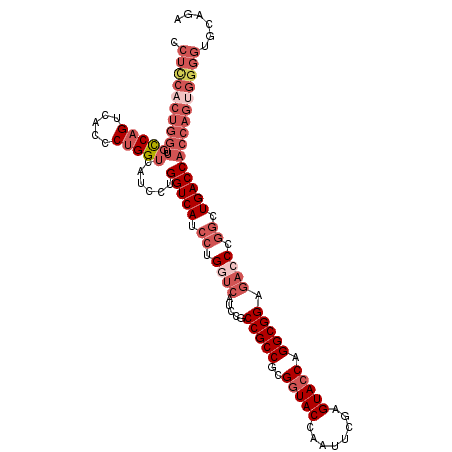
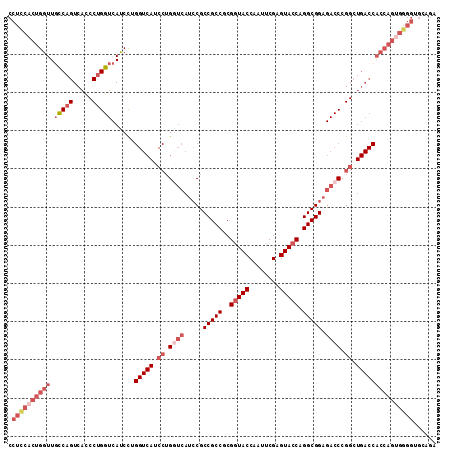
| Location | 20,430,710 – 20,430,822 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -52.50 |
| Consensus MFE | -39.19 |
| Energy contribution | -42.68 |
| Covariance contribution | 3.49 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20430710 112 - 22407834 UCUGCACCCCACUGGUGGUCAGCCGGGUCUCCGCCUGGUACUCGAAUUGGUACCGCGGCGGCGGAUGAUCAGGAUGACCAGGAUGACCAGGGUGACUGGAAACCAGUGGAGG ......(((((((((((((((.((.(((((((((((((((((......)))))))....)))))).)))).)).))))).......((((.....))))..)))))))).)) ( -57.30) >DroSec_CAF1 2105 112 - 1 UCUGCACCCCACUGGUGGUCAGCCGGGUCUCCGCCUGGUACUCGAACUGGUACCGCGGCGGCGGAUGACCAGGAUGACCAGGGUGACCAGGGUGACUGGCAACCAGUGGAGG ......(((((((((((((((.((.(((((((((((((((((......)))))))....)))))).)))).)).))))).((....)).((....))....)))))))).)) ( -61.00) >DroSim_CAF1 3039 104 - 1 NNNNNNNNNNNNNNNNNNNNNNCCGGGUCUCCGCCUGGUACUCGAAUUGGUACCGCGGCGGCGCGUCUUCA--------AGGGUGACCAGGGUGACUGGCAACCAGUGGAGG .......................((.(((.((((..((((((......)))))))))).))).)).(((((--------..(((..((((.....))))..)))..))))). ( -38.60) >DroEre_CAF1 1966 112 - 1 UCUGCACCCCACUGGUGGUCAGCCGGGUCUCCGCCUGGUACUCGAAUUGGUACCGCGGCGGCGGAUGACCAGGAUGACCCAGAUGACCAGGAUGACUGACAACCAGGGAAGG ........((.((((((((((.((.(((((((((((((((((......)))))))....)))))).)))).)).)))))........(((.....)))...))))))).... ( -48.80) >DroYak_CAF1 3134 112 - 1 UCUGCACCCCACUGGUGGUCAGCCGGGUCUCCGCCUGGUACUCGAAUUGGUAUCGCGGCGGCGGAUGACCUGGAUGACCUAGAUGACCUGGAUGACUGGCAACCAGUGGAGG ......(((((((((((((((.((((((((((((((((((((......)))))))....)))))).))))))).)))))....((.((.(.....).)))))))))))).)) ( -56.80) >consensus UCUGCACCCCACUGGUGGUCAGCCGGGUCUCCGCCUGGUACUCGAAUUGGUACCGCGGCGGCGGAUGACCAGGAUGACCAGGAUGACCAGGGUGACUGGCAACCAGUGGAGG ......(((((((((((((((.((.(((((((((((((((((......)))))))....)))))).)))).)).))))).......((((.....))))..)))))))).)) (-39.19 = -42.68 + 3.49)

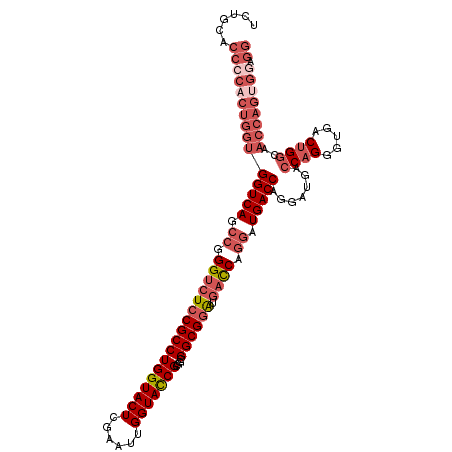
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:32:31 2006