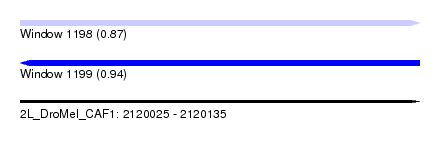
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,120,025 – 2,120,135 |
| Length | 110 |
| Max. P | 0.937570 |

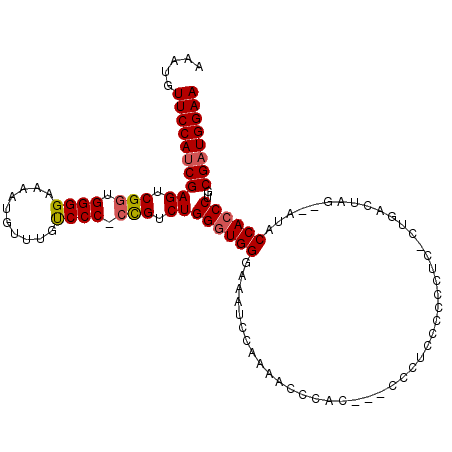
| Location | 2,120,025 – 2,120,135 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.15 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -29.23 |
| Energy contribution | -29.16 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2120025 110 + 22407834 UUCCAUCGAAGGGUGGUAUCUCUAGUCAG-CAGGGGGGAGGGUGGUUGGGUUUUGAAUUUCCCACCCAGACGG-GGGACAAACAUUUUCCCCACCGACUCGAUGGAACAUUU (((((((((((((......)))).(((..-..((((((((((((..((((..........))))(((....))-).......)))))))))).))))))))))))))..... ( -43.10) >DroSec_CAF1 120372 105 + 1 UUCCAUCGCAGGGUGGUAU--CUAGUCAC-GAGGGGGGAGGG---GUGGUUUUUGGAUUUCCCACCCAGACGG-GGGACAAACAUUUUCCCCACCGACUCGAUGGAACAUUU ((((((((.((..((((..--...(((..-..(((((((((.---............)))))).))).)))((-((((........)))))))))).))))))))))..... ( -39.72) >DroSim_CAF1 120158 105 + 1 UUCCAUCGCAGGGUGGUAU--CUAGUCAG-GAGGGGGGAGGG---AUGGGUUUUGGAUUUCCCAACCAGACGG-GGGAGAAACAUUUUCCCCACCGACUCGAUGGAACAUUU ((((((((.((..((((..--...(((.(-(..(((..(..(---(......))...)..)))..)).)))((-((((((....)))))))))))).))))))))))..... ( -41.40) >DroYak_CAF1 125338 110 + 1 UUCCAACGCAGGGUGGUGU--GUAGCCAGAGUGGGGGGAGGGGAAGAGGGGUUUAGAUUUUCCACCCAGACAGGGGGGUAAACAUUUUCCCCAUCGACUCGAUGGAACAUUU (((((.(......((((..--...))))((((.((....((((((((...(((((.....(((.(((.....))))))))))).)))))))).)).))))).)))))..... ( -37.80) >consensus UUCCAUCGCAGGGUGGUAU__CUAGUCAG_GAGGGGGGAGGG___GUGGGUUUUGGAUUUCCCACCCAGACGG_GGGACAAACAUUUUCCCCACCGACUCGAUGGAACAUUU ((((((((.(((((((................(((((((((................)))))).))).......((((........)))))))))..))))))))))..... (-29.23 = -29.16 + -0.06)

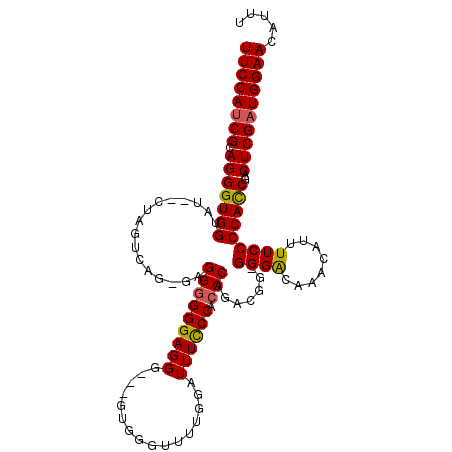
| Location | 2,120,025 – 2,120,135 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.15 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -27.23 |
| Energy contribution | -27.61 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2120025 110 - 22407834 AAAUGUUCCAUCGAGUCGGUGGGGAAAAUGUUUGUCCC-CCGUCUGGGUGGGAAAUUCAAAACCCAACCACCCUCCCCCCUG-CUGACUAGAGAUACCACCCUUCGAUGGAA .....((((((((((((((((((((.........))))-).....((((((................))))))........)-)))))).(((........))))))))))) ( -36.99) >DroSec_CAF1 120372 105 - 1 AAAUGUUCCAUCGAGUCGGUGGGGAAAAUGUUUGUCCC-CCGUCUGGGUGGGAAAUCCAAAAACCAC---CCCUCCCCCCUC-GUGACUAG--AUACCACCCUGCGAUGGAA .....((((((((....((((((((.........))).-..(((.(((.((((..............---...)))))))..-..)))...--...)))))...)))))))) ( -33.93) >DroSim_CAF1 120158 105 - 1 AAAUGUUCCAUCGAGUCGGUGGGGAAAAUGUUUCUCCC-CCGUCUGGUUGGGAAAUCCAAAACCCAU---CCCUCCCCCCUC-CUGACUAG--AUACCACCCUGCGAUGGAA .....((((((((....((((((((.........))).-..(((((((..(((..............---..........))-)..)))))--)).)))))...)))))))) ( -35.36) >DroYak_CAF1 125338 110 - 1 AAAUGUUCCAUCGAGUCGAUGGGGAAAAUGUUUACCCCCCUGUCUGGGUGGAAAAUCUAAACCCCUCUUCCCCUCCCCCCACUCUGGCUAC--ACACCACCCUGCGUUGGAA .....(((((.((((..((.((((.....((((.((.(((.....))).)).)))).....))))...))..))).........(((....--...)))......).))))) ( -25.80) >consensus AAAUGUUCCAUCGAGUCGGUGGGGAAAAUGUUUGUCCC_CCGUCUGGGUGGGAAAUCCAAAACCCAC___CCCUCCCCCCUC_CUGACUAG__AUACCACCCUGCGAUGGAA .....((((((((((.(((.((((..........)))).))).))((((((.............................................))))))..)))))))) (-27.23 = -27.61 + 0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:08 2006